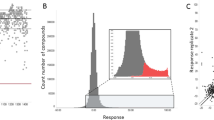
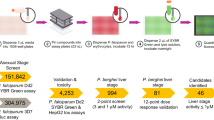
Abstract
Malaria is a devastating infection caused by protozoa of the genus Plasmodium. Drug resistance is widespread, no new chemical class of antimalarials has been introduced into clinical practice since 1996 and there is a recent rise of parasite strains with reduced sensitivity to the newest drugs. We screened nearly 2 million compounds in GlaxoSmithKline’s chemical library for inhibitors of P. falciparum, of which 13,533 were confirmed to inhibit parasite growth by at least 80% at 2 µM concentration. More than 8,000 also showed potent activity against the multidrug resistant strain Dd2. Most (82%) compounds originate from internal company projects and are new to the malaria community. Analyses using historic assay data suggest several novel mechanisms of antimalarial action, such as inhibition of protein kinases and host–pathogen interaction related targets. Chemical structures and associated data are hereby made public to encourage additional drug lead identification efforts and further research into this disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
World Health Organization. World malaria report. 〈http://www.who.int/malaria/publications/atoz/9789241563901/en/index.html〉 (2009)
Anstey, N. M., Russell, B., Yeo, T. W. & Price, R. N. The pathophysiology of vivax malaria. Trends Parasitol. 25, 220–227 (2009)
Ekland, E. H. & Fidock, D. A. In vitro evaluations of antimalarial drugs and their relevance to clinical outcomes. Int. J. Parasitol. 38, 743–747 (2008)
Andriantsoanirina, V. et al. Plasmodium falciparum drug resistance in Madagascar: facing the spread of unusual pfdhfr and pfmdr-1 haplotypes and the decrease of dihydroartemisinin susceptibility. Antimicrob. Agents Chemother. 53, 4588–4597 (2009)
Bonnet, M. et al. Varying efficacy of artesunate+amodiaquine and artesunate+sulphadoxine-pyrimethamine for the treatment of uncomplicated falciparum malaria in the Democratic Republic of Congo: a report of two in-vivo studies. Malar. J. 8, 192 (2009)
Carrara, V. I. et al. Changes in the treatment responses to artesunate-mefloquine on the northwestern border of Thailand during 13 years of continuous deployment. PLoS ONE 4, e4551 (2009)
Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nature Rev. Drug Discov. 6, 29–40 (2007)
O'Brien, P. J. et al. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch. Toxicol. 80, 580–604 (2006)
Xia, M. et al. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ. Health Perspect. 116, 284–291 (2008)
Yuan, J. et al. Genetic mapping of targets mediating differential chemical phenotypes in Plasmodium falciparum . Nature Chem. Biol. 5, 765–771 (2009)
Reed, M. B., Saliba, K. J., Caruana, S. R., Kirk, K. & Cowman, A. F. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum . Nature 403, 906–909 (2000)
Bemis, G. W. & Murcko, M. A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 39, 2887–2893 (1996)
Daylight Chemical Information Systems, Inc. Daylight theory manual. 〈http://www.daylight.com/dayhtml/doc/theory/index.html〉 (2008)
Johnson, M., Lajiness, M. & Maggiora, G. M. in Qsar: Quantitative structure-activity relationships in drug design (ed Fauchere, J. L.) 167–171 (Alan R. Liss, 1989)
Davis, A. M., Keeling, D. J., Steele, J., Tomkinson, N. P. & Tinker, A. C. Components of successful lead generation. Curr. Top. Med. Chem. 5, 421–439 (2005)
Köhler, S. et al. A plastid of probable green algal origin in apicomplexan parasites. Science 275, 1485–1489 (1997)
McFadden, G. I., Reith, M. E., Munholland, J. & Lang-Unnasch, N. Plastid in human parasites. Nature 381, 482 (1996)
Le Roch, K. G. et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508 (2003)
Bozdech, Z. et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum . PLoS Biol. 1, e5 (2003)
Wu, Y., Wang, X., Liu, X. & Wang, Y. Data-mining approaches reveal hidden families of proteases in the genome of malaria parasite. Genome Res. 13, 601–616 (2003)
Leroy, D. & Doerig, C. Drugging the Plasmodium kinome: The benefits of academia–industry synergy. Trends Pharmacol. Sci. 29, 241–249 (2008)
Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002)
Anamika, S. N. & Krupa, A. A genomic perspective of protein kinases in Plasmodium falciparum . Proteins 58, 180–189 (2005)
Ward, P., Equinet, L., Packer, J. & Doerig, C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics 5, 79 (2004)
Karaman, M. W. et al. A quantitative analysis of kinase inhibitor selectivity. Nature Biotechnol. 26, 127–132 (2008)
Madeira, L. et al. Genome-wide detection of serpentine receptor-like proteins in malaria parasites. PLoS ONE 3, e1889 (2008)
Aurrecoechea, C. et al. PlasmoDB: A functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543 (2009)
Trager, W. & Jensen, J. B. Human malaria parasites in continuous culture. Science 193, 673–675 (1976)
Makler, M. T. & Hinrichs, D. J. Measurement of the lactate dehydrogenase activity of Plasmodium falciparum as an assessment of parasitemia. Am. J. Trop. Med. Hyg. 48, 205–210 (1993)
Zhang, J. H., Chung, T. D. Y. & Oldenburg, K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999)
Desjardins, R. E., Canfield, C. J., Haynes, J. D. & Chulay, J. D. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother. 16, 710–718 (1979)
Goodman, C. D., Su, V. & McFadden, G. I. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum . Mol. Biochem. Parasitol. 152, 181–191 (2007)
Ramya, T. N. C., Mishra, S., Karmodiya, K., Surolia, N. & Surolia, A. Inhibitors of nonhousekeeping functions of the apicoplast defy delayed death in Plasmodium falciparum . Antimicrob. Agents Chemother. 51, 307–316 (2007)
Fidock, D. A., Rosenthal, P. J., Croft, S. L., Brun, R. & Nwaka, S. Antimalarial drug discovery: Efficacy models for compound screening. Nature Rev. Drug Discov. 3, 509–520 (2004)
Aureus Pharma. AurSCOPE database. 〈http://www.aureus-pharma.com/Pages/Products/Aurscope.php〉 (2004)
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994)
Felsentein, J. Phylip (phylogenetic inference package) v3.67. Department of Genetics, University of Washington 〈http://evolution.genetics.washington.edu/phylip.html〉.
Huson, D. H. et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8, 460 (2007)
Acknowledgements
We thank S. Peregrina and S. Prats for technical assistance, and D. Jiménez-Alfaro for supplying compounds pre-dispensed in microtitre plates. We thank P. Vallance and R. Keenan for organising support for this work, R. Macarron and J. Luengo for developing the IFI index and getting the chemistry data ready for publication, together with J. M. Fiandor, S. Chakravorty and members of GSK’s Chemistry Council. J. Lewis, A. Clow, J. Overington and M. Davies were instrumental in the uploading and formatting of the data. We also thank N. Cammack, P. Sanseau and J. Burrows for critically commenting on the manuscript. The support and funding of Medicines for Malaria Venture is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
F.-J.G. and J.F.G.-B. planned and designed the work. F.-J.G. supervised all experimental work and analysed the screening data, L.M.S., J.V., C.d.C. and E.A. performed the screening assays and contributed to data analysis. J.-L.L., D.E.V., D.V.S.G. and C.E.P. performed the cheminformatic analyses and wrote sections of the manuscript. V.K., S.H. and J.R.B. performed the bioinformatic analyses and J.R.B. contributed the relevant sections to the manuscript. L.R.C and J.F.G.-B integrated individual contributions and wrote the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Chemical structures and data described have been deposited at EBI (http://www.ebi.ac.uk/chemblntd).
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-3 with legends and Supplementary Tables 3-5. (PDF 675 kb)
Supplementary Table 1
This table contains an annotated list of all positives from the P. falciparum screen. Structures are shown as "SMILES" codes. (XLS 3451 kb)
Supplementary Table 2
This table shows compounds in TCAMS with literature references on their mode of action. (XLS 437 kb)
Supplementary Data 1
The file contains the multiple kinase sequence alignment. This file was added on 2 June 2010. (TXT 173 kb)
Rights and permissions
About this article
Cite this article
Gamo, FJ., Sanz, L., Vidal, J. et al. Thousands of chemical starting points for antimalarial lead identification. Nature 465, 305–310 (2010). https://doi.org/10.1038/nature09107
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09107
This article is cited by
-
Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase
Nature Communications (2024)
-
Isoliensinine from Cissampelos pariera rhizomes exhibits potential gametocytocidal and anti-malarial activities against Plasmodium falciparum clinical isolates
Malaria Journal (2023)
-
Enhancing drug property prediction with dual-channel transfer learning based on molecular fragment
BMC Bioinformatics (2023)
-
Identifying inhibitors of β-haematin formation with activity against chloroquine-resistant Plasmodium falciparum malaria parasites via virtual screening approaches
Scientific Reports (2023)
-
Antimalarial drug discovery: progress and approaches
Nature Reviews Drug Discovery (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.