Abstract
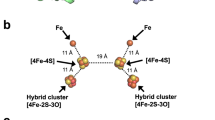
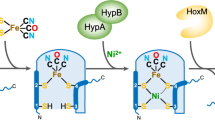
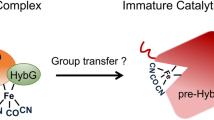
Complex enzymes containing Fe–S clusters are ubiquitous in nature, where they are involved in a number of fundamental processes including carbon dioxide fixation, nitrogen fixation and hydrogen metabolism1,2. Hydrogen metabolism is facilitated by the activity of three evolutionarily and structurally unrelated enzymes: the [NiFe]-hydrogenases, [FeFe]-hydrogenases and [Fe]-hydrogenases3,4 (Hmd). The catalytic core of the [FeFe]-hydrogenase (HydA), termed the H-cluster, exists as a [4Fe–4S] subcluster linked by a cysteine thiolate to a modified 2Fe subcluster with unique non-protein ligands5,6. The 2Fe subcluster and non-protein ligands are synthesized by the hydrogenase maturation enzymes HydE, HydF and HydG; however, the mechanism, synthesis and means of insertion of H-cluster components remain unclear7,8,9,10. Here we show the structure of HydAΔEFG (HydA expressed in a genetic background devoid of the active site H-cluster biosynthetic genes hydE, hydF and hydG) revealing the presence of a [4Fe–4S] cluster and an open pocket for the 2Fe subcluster. The structure indicates that H-cluster synthesis occurs in a stepwise manner, first with synthesis and insertion of the [4Fe–4S] subcluster by generalized host-cell machinery11,12 and then with synthesis and insertion of the 2Fe subcluster by specialized hydE-, hydF- and hydG-encoded maturation machinery7,8,9,10. Insertion of the 2Fe subcluster presumably occurs through a cationically charged channel that collapses following incorporation, as a result of conformational changes in two conserved loop regions. The structure, together with phylogenetic analysis, indicates that HydA emerged within bacteria most likely from a Nar1-like ancestor lacking the 2Fe subcluster, and that this was followed by acquisition in several unicellular eukaryotes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Drennan, C. L. & Peters, J. W. Surprising cofactors in metalloenzymes. Curr. Opin. Struct. Biol. 13, 220–226 (2003)
Fontecilla-Camps, J. C., Amara, P., Cavazza, C., Nicolet, Y. & Volbeda, A. Structure-function relationships of anaerobic gas-processing metalloenzymes. Nature 460, 814–822 (2009)
Vignais, P. M. & Billoud, B. Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272 (2007)
Shima, S. & Thauer, R. K. A third type of hydrogenase catalyzing H2 activation. Chem. Rec. 7, 37–46 (2007)
Peters, J. W., Lanzilotta, W. N., Lemon, B. J. & Seefeldt, L. C. X-ray crystal structure of the Fe-only hydrogenase (Cpl) from Clostridium pasteurianum to 1.8 angstrom resolution. Science 282, 1853–1858 (1998)
Nicolet, Y., Piras, C., Legrand, P., Hatchikian, C. E. & Fontecilla-Camps, J. C. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure 7, 13–23 (1999)
Nicolet, Y. et al. X-ray structure of the [FeFe]-hydrogenase maturase HydE from Thermotoga maritima . J. Biol. Chem. 283, 18861–18872 (2008)
McGlynn, S. E. et al. HydF as a scaffold protein in [FeFe] hydrogenase H-cluster biosynthesis. FEBS Lett. 582, 2183–2187 (2008)
Pilet, E. et al. The role of the maturase HydG in [FeFe]-hydrogenase active site synthesis and assembly. FEBS Lett. 583, 506–511 (2009)
Mulder, D. W. et al. Activation of HydAΔEFG requires a preformed [4Fe-4S] cluster. Biochemistry 48, 6240–6248 (2009)
Lill, R. Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 (2009)
Johnson, D. C., Dean, D. R., Smith, A. D. & Johnson, M. K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281 (2005)
Schwarz, G., Mendel, R. R. & Ribbe, M. W. Molybdenum cofactors, enzymes and pathways. Nature 460, 839–847 (2009)
Posewitz, M. C. et al. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J. Biol. Chem. 279, 25711–25720 (2004)
Rubach, J. K., Brazzolotto, X., Gaillard, J. & Fontecave, M. Biochemical characterization of the HydE and HydG iron-only hydrogenase maturation enzymes from Thermatoga maritima . FEBS Lett. 579, 5055–5060 (2005)
Brazzolotto, X. et al. The [Fe-Fe]-hydrogenase maturation protein HydF from Thermotoga maritima is a GTPase with an iron-sulfur cluster. J. Biol. Chem. 281, 769–774 (2006)
McGlynn, S. E., Mulder, D. W., Shepard, E. M., Broderick, J. B. & Peters, J. W. Hydrogenase cluster biosynthesis: organometallic chemistry nature’s way. Dalton Trans. 22, 4274–4285 (2009)
Peters, J. W., Szilagyi, R. K., Naumov, A. & Douglas, T. A radical solution for the biosynthesis of the H-cluster of hydrogenase. FEBS Lett. 580, 363–367 (2006)
Balk, J., Pierik, A. J., Netz, D. J., Muhlenhoff, U. & Lill, R. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23, 2105–2115 (2004)
Peters, J. W. et al. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry 36, 1181–1187 (1997)
Einsle, O. et al. Nitrogenase MoFe-protein at 1.16 Å resolution: a central ligand in the FeMo-cofactor. Science 297, 1696–1700 (2002)
Schmid, B. et al. Structure of a cofactor-deficient nitrogenase MoFe protein. Science 296, 352–356 (2002)
Fani, R., Gallo, R. & Lio, P. Molecular evolution of nitrogen fixation: the evolutionary history of the nifD, nifK, nifE, and nifN genes. J. Mol. Evol. 51, 1–11 (2000)
Meyer, J. [FeFe] hydrogenases and their evolution: a genomic perspective. Cell. Mol. Life Sci. 64, 1063–1084 (2007)
Hug, L. A., Stechmann, A. & Roger, A. J. Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes. Mol. Biol. Evol. 27, 311–324 (2010)
Rubio, L. M. & Ludden, P. W. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 62, 93–111 (2008)
Georgiadis, M. M. et al. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii . Science 257, 1653–1659 (1992)
DeLano, W. L. PyMOL Molecular Viewer 〈http://www.pymol.org〉 (2002)
Pandey, A. S., Harris, T. V., Giles, L. J., Peters, J. W. & Szilagyi, R. K. Dithiomethylether as a ligand in the hydrogenase H-cluster. J. Am. Chem. Soc. 130, 4533–4540 (2008)
Silakov, A., Wenk, B., Reijerse, E. & Lubitz, W. 14N HYSCORE investigation of the H-cluster of [FeFe] hydrogenase: evidence for a nitrogen in the dithiol bridge. Phys. Chem. Chem. Phys. 11, 6592–6599 (2009)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative. Computation Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Perrakis, A., Morris, R. & Lamzin, V. S. Automated protein model building combined with iterative structure refinement. Nature Struct. Biol. 6, 458–463 (1999)
Terwilliger, T. C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
Ten Eyck, L. F. Crystallographic fast Fourier transforms. Acta Crystallogr. A 29, 183–191 (1973)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003)
Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 (2008)
Abascal, F., Zardoya, R. & Posada, D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005)
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003)
Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001)
Acknowledgements
This work was supported by a US Air Force Office of Scientific Research Multidisciplinary University Research Initiative Award (FA9550-05-01-0365, J.W.P.) and the NASA Astrobiology Institute (NAI)-funded Astrobiology Biogeocatalysis Research Center (NNA08C-N85A, J.B.B. and J.W.P.). E.S.B. was supported by a NAI postdoctoral fellowship. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology programme is supported by the US Department of Energy, Office of Biological and Environmental Research, the US National Institutes of Health, National Center for Research Resources, Biomedical Technology programme, and the US National Institute of General Medical Sciences.
Author information
Authors and Affiliations
Contributions
Author Contributions The structural work was conducted by D.W.M. with contributions from R.S. and J.A.E. E.S.B. led the phylogenetic work with contributions from R.K.L. J.W.P. supervised the work with assistance from J.B.B. D.W.M., E.S.B. and J.W.P. led the manuscript preparation with contributions from J.B.B., R.S., R.K.L. and J.A.E.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
This file contains Supplementary Figures 1-5 with legends, Supplementary Tables 1-2, a Supplementary Discussion and References. (PDF 7114 kb)
Rights and permissions
About this article
Cite this article
Mulder, D., Boyd, E., Sarma, R. et al. Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydAΔEFG. Nature 465, 248–251 (2010). https://doi.org/10.1038/nature08993
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08993
This article is cited by
-
A personal account on 25 years of scientific literature on [FeFe]-hydrogenase
JBIC Journal of Biological Inorganic Chemistry (2023)
-
Enzyme-like water preorganization in a synthetic molecular cleft for homogeneous water oxidation catalysis
Nature Catalysis (2022)
-
Stability of the H-cluster under whole-cell conditions—formation of an Htrans-like state and its reactivity towards oxygen
JBIC Journal of Biological Inorganic Chemistry (2022)
-
Where do the electrons go? How numerous redox processes drive phytochemical diversity
Phytochemistry Reviews (2021)
-
Bioassembly of complex iron–sulfur enzymes: hydrogenases and nitrogenases
Nature Reviews Chemistry (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.