Abstract
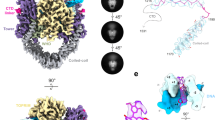
Type II topoisomerases are required for the management of DNA tangles and supercoils1, and are targets of clinical antibiotics and anti-cancer agents2. These enzymes catalyse the ATP-dependent passage of one DNA duplex (the transport or T-segment) through a transient, double-stranded break in another (the gate or G-segment), navigating DNA through the protein using a set of dissociable internal interfaces, or ‘gates’3,4. For more than 20 years, it has been established that a pair of dimer-related tyrosines, together with divalent cations, catalyse G-segment cleavage5,6,7. Recent efforts have proposed that strand scission relies on a ‘two-metal mechanism’8,9,10, a ubiquitous biochemical strategy that supports vital cellular processes ranging from DNA synthesis to RNA self-splicing11,12. Here we present the structure of the DNA-binding and cleavage core of Saccharomyces cerevisiae topoisomerase II covalently linked to DNA through its active-site tyrosine at 2.5 Å resolution, revealing for the first time the organization of a cleavage-competent type II topoisomerase configuration. Unexpectedly, metal-soaking experiments indicate that cleavage is catalysed by a novel variation of the classic two-metal approach. Comparative analyses extend this scheme to explain how distantly-related type IA topoisomerases cleave single-stranded DNA, unifying the cleavage mechanisms for these two essential enzyme families. The structure also highlights a hitherto undiscovered allosteric relay that actuates a molecular ‘trapdoor’ to prevent subunit dissociation during cleavage. This connection illustrates how an indispensable chromosome-disentangling machine auto-regulates DNA breakage to prevent the aberrant formation of mutagenic and cytotoxic genomic lesions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
03 June 2010
A correction was made to Fig. 1a.
References
Schoeffler, A. J. & Berger, J. M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41, 41–101 (2008)
Deweese, J. E. & Osheroff, N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res. 37, 738–748 (2009)
Roca, J. & Wang, J. C. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell 71, 833–840 (1992)
Roca, J., Berger, J. M., Harrison, S. C. & Wang, J. C. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA 93, 4057–4062 (1996)
Worland, S. T. & Wang, J. C. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae . J. Biol. Chem. 264, 4412–4416 (1989)
Goto, T. & Wang, J. C. Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. J. Biol. Chem. 257, 5866–5872 (1982)
Goto, T., Laipis, P. & Wang, J. C. The purification and characterization of DNA topoisomerases I and II of the yeast Saccharomyces cerevisiae . J. Biol. Chem. 259, 10422–10429 (1984)
West, K. L. et al. Mutagenesis of E477 or K505 in the B′ domain of human topoisomerase IIβ increases the requirement for magnesium ions during strand passage. Biochemistry 39, 1223–1233 (2000)
Noble, C. G. & Maxwell, A. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J. Mol. Biol. 318, 361–371 (2002)
Deweese, J. E., Guengerich, F. P., Burgin, A. B. & Osheroff, N. Metal ion interactions in the DNA cleavage/ligation active site of human topoisomerase IIα. Biochemistry 48, 8940–8947 (2009)
Steitz, T. A. & Steitz, J. A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA 90, 6498–6502 (1993)
Yang, W., Lee, J. Y. & Nowotny, M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006)
Dong, K. C. & Berger, J. M. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature 450, 1201–1205 (2007)
Burgin, A. B. Jr, Huizenga, B. N. & Nash, H. A. A novel suicide substrate for DNA topoisomerases and site-specific recombinases. Nucleic Acids Res. 23, 2973–2979 (1995)
Redinbo, M. R., Stewart, L., Kuhn, P., Champoux, J. J. & Hol, W. G. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 279, 1504–1513 (1998)
Wang, J., Yu, P., Lin, T. C., Konigsberg, W. H. & Steitz, T. A. Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions. Biochemistry 35, 8110–8119 (1996)
Nowotny, M. & Yang, W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 25, 1924–1933 (2006)
Aravind, L., Leipe, D. D. & Koonin, E. V. Toprim–a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26, 4205–4213 (1998)
Liu, Q. & Wang, J. C. Identification of active site residues in the “GyrA” half of yeast DNA topoisomerase II. J. Biol. Chem. 273, 20252–20260 (1998)
Okada, Y. et al. Assignment of functional amino acids around the active site of human DNA topoisomerase IIα. J. Biol. Chem. 275, 24630–24638 (2000)
Nichols, M. D., DeAngelis, K., Keck, J. L. & Berger, J. M. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 18, 6177–6188 (1999)
Berger, J. M., Fass, D., Wang, J. C. & Harrison, S. C. Structural similarities between topoisomerases that cleave one or both DNA strands. Proc. Natl Acad. Sci. USA 95, 7876–7881 (1998)
Domanico, P. L. & Tse-Dinh, Y. C. Mechanistic studies on E. coli DNA topoisomerase I: divalent ion effects. J. Inorg. Biochem. 42, 87–96 (1991)
Changela, A., DiGate, R. J. & Mondragon, A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature 411, 1077–1081 (2001)
Strahs, D., Zhu, C. X., Cheng, B., Chen, J. & Tse-Dinh, Y. C. Experimental and computational investigations of Ser10 and Lys13 in the binding and cleavage of DNA substrates by Escherichia coli DNA topoisomerase I. Nucleic Acids Res. 34, 1785–1797 (2006)
Cheng, B. et al. Asp-to-Asn substitution at the first position of the DxD TOPRIM motif of recombinant bacterial topoisomerase I is extremely lethal to E. coli . J. Mol. Biol. 385, 558–567 (2009)
Berger, J. M., Gamblin, S. J., Harrison, S. C. & Wang, J. C. Structure and mechanism of DNA topoisomerase II. Nature 379, 225–232 (1996)
Morais Cabral, J. H. et al. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388, 903–906 (1997)
Fass, D., Bogden, C. E. & Berger, J. M. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nature Struct. Biol. 6, 322–326 (1999)
Deweese, J. E., Burgin, A. B. & Osheroff, N. Using 3′-bridging phosphorothiolates to isolate the forward DNA cleavage reaction of human topoisomerase IIα. Biochemistry 47, 4129–4140 (2008)
Tropea, J. E., Cherry, S. & Waugh, D. S. Expression and purification of soluble His6-tagged TEV protease. Methods Mol. Biol. 498, 297–307 (2009)
Sabbagh, G., Fettes, K. J., Gosain, R., O’Neil, I. A. & Cosstick, R. Synthesis of phosphorothioamidites derived from 3′-thio-3′-deoxythymidine and 3′-thio-2′,3′-dideoxycytidine and the automated synthesis of oligodeoxynucleotides containing a 3′-S-phosphorothiolate linkage. Nucleic Acids Res. 32, 495–501 (2004)
MacDowell, A. A. et al. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J. Synchrotron Radiat. 11, 447–455 (2004)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004)
DeLano, W. L. The PyMOL Molecular Graphics System. (http://www.pymol.org) (2002)
Chen, X. et al. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93, 827–839 (1998)
Zechiedrich, E. L., Christiansen, K., Andersen, A. H., Westergaard, O. & Osheroff, N. Double-stranded DNA cleavage/religation reaction of eukaryotic topoisomerase II: evidence for a nicked DNA intermediate. Biochemistry 28, 6229–6236 (1989)
Acknowledgements
This work was supported the NIH (GM033944 and GM053960 for N.O.; T32CA09592 for J.E.D.; GM08295 for B.H.S. and CA077373 for J.M.B.).
Author information
Authors and Affiliations
Contributions
B.H.S. purified the complex, grew the crystals, and solved the structures. J.M.B. assisted with refinement and inspection of the molecular models. A.B.B. synthesized the phosphorothiolate reagent. J.E.D. assisted the design of the DNA substrate and optimizing cleavage conditions. N.O. and J.M.B. designed the experiments. All authors contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table S1, Supplementary Figures S1-S6 with legends, Supplementary Movie SM1 legend and References. (PDF 929 kb)
Supplementary Movie SM1
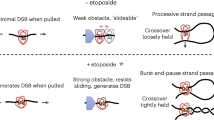
The movie shows that the transition between cleaved and uncleaved DNA states permits C-gate opening (see Supplementary Information file for full legend). (MOV 19109 kb)
Rights and permissions
About this article
Cite this article
Schmidt, B., Burgin, A., Deweese, J. et al. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature 465, 641–644 (2010). https://doi.org/10.1038/nature08974
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08974
This article is cited by
-
Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin
Nature Catalysis (2023)
-
Etoposide promotes DNA loop trapping and barrier formation by topoisomerase II
Nature Chemical Biology (2023)
-
Structural and biochemical basis for DNA and RNA catalysis by human Topoisomerase 3β
Nature Communications (2022)
-
Potent DNA gyrase inhibitors bind asymmetrically to their target using symmetrical bifurcated halogen bonds
Nature Communications (2021)
-
The Prevailing Role of Topoisomerase 2 Beta and its Associated Genes in Neurons
Molecular Neurobiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.