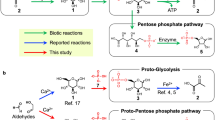
Abstract
Most archaeal groups and deeply branching bacterial lineages harbour thermophilic organisms with a chemolithoautotrophic metabolism. They live at high temperatures in volcanic habitats at the expense of inorganic substances, often under anoxic conditions1. These autotrophic organisms use diverse carbon dioxide fixation mechanisms generating acetyl-coenzyme A, from which gluconeogenesis must start2,3,4. Here we show that virtually all archaeal groups as well as the deeply branching bacterial lineages contain a bifunctional fructose 1,6-bisphosphate (FBP) aldolase/phosphatase with both FBP aldolase and FBP phosphatase activity. This enzyme is missing in most other Bacteria and in Eukaryota, and is heat-stabile even in mesophilic marine Crenarchaeota. Its bifunctionality ensures that heat-labile triosephosphates are quickly removed and trapped in stabile fructose 6-phosphate, rendering gluconeogenesis unidirectional. We propose that this highly conserved, heat-stabile and bifunctional FBP aldolase/phosphatase represents the pace-making ancestral gluconeogenic enzyme, and that in evolution gluconeogenesis preceded glycolysis5.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Stetter, K. O. Hyperthermophiles in the history of life. Phil. Trans. R. Soc. Lond. B 361, 1837–1843 (2006)
Berg, I. A. et al. Autotrophic carbon fixation in Archaea. Nature Rev. Microbiol. (in the press)
Huber, H. et al. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis . Proc. Natl Acad. Sci. USA 105, 7851–7856 (2008)
Berg, I. A., Kockelkorn, D., Buckel, W. & Fuchs, G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318, 1782–1786 (2007)
Ronimus, R. S. & Morgan, H. W. Distribution and phylogenies of enzymes of the Embden-Meyerhof-Parnas pathway from archaea and hyperthermophilic bacteria support a gluconeogenic origin of metabolism. Archaea 1, 199–221 (2003)
Wächtershäuser, G. On the chemistry and evolution of the pioneer organism. Chem. Biodivers. 4, 584–602 (2007)
Martin, W., Baross, J., Kelley, D. & Russell, M. J. Hydrothermal vents and the origin of life. Nature Rev. Microbiol. 6, 805–814 (2008)
Siebers, B. & Schönheit, P. Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr. Opin. Microbiol. 8, 695–705 (2005)
Fuchs, G., Winter, H., Steiner, I. & Stupperich, E. Enzymes of gluconeogenesis in the autotroph Methanobacterium thermoautotrophicum . Arch. Microbiol. 136, 160–162 (1983)
Jahn, U., Huber, H., Eisenreich, W., Hügler, M. & Fuchs, G. Insights into the autotrophic CO2 fixation pathway of the archaeon Ignicoccus hospitalis: comprehensive analysis of the central carbon metabolism. J. Bacteriol. 189, 4108–4119 (2007)
Schäfer, S., Barkowski, C. & Fuchs, G. Carbon assimilation by the autotrophic thermophilic archaebacterium Thermoproteus neutrophilus . Arch. Microbiol. 146, 301–308 (1986)
Imanaka, H., Fukui, T., Atomi, H. & Imanaka, T. Gene cloning and characterization of fructose-1,6-bisphosphate aldolase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biosci. Bioeng. 94, 237–243 (2002)
Lorentzen, E., Siebers, B., Hensel, R. & Pohl, E. Structure, function and evolution of the Archaeal class I fructose-1,6-bisphosphate aldolase. Biochem. Soc. Trans. 32, 259–263 (2004)
Andreeva, A. et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 36, D419–D425 (2008)
Grochowski, L. L. & White, R. H. Promiscuous anaerobes: new and unconventional metabolism in methanogenic archaea. Ann. NY Acad. Sci. 1125, 190–214 (2008)
Rashid, N. et al. A novel candidate for the true fructose-1,6-bisphosphatase in archaea. J. Biol. Chem. 277, 30649–30655 (2002)
Sato, T. et al. Genetic evidence identifying the true gluconeogenic fructose-1,6-bisphosphatase in Thermococcus kodakaraensis and other hyperthermophiles. J. Bacteriol. 186, 5799–5807 (2004)
Nishimasu, H., Fushinobu, S., Shoun, H. & Wakagi, T. The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea. Structure 12, 949–959 (2004)
Hallam, S. J. et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum . Proc. Natl Acad. Sci. USA 103, 18296–18301 (2006)
Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Potential genomic determinants of hyperthermophily. Trends Genet. 19, 172–176 (2003)
Chong, P. K., Burja, A. M., Radianingtyas, H., Fazeli, A. & Wright, P. C. Proteome analysis of Sulfolobus solfataricus P2 propanol metabolism. J. Proteome Res. 6, 1430–1439 (2007)
Ludwig, W. & Klenk, H.-P. In The Archaea and the Deeply Branching and Phototrophic Bacteria (eds Boone, D. R. & Garrity, G. M.). Bergey’s Manual of Systematic Bacteriology, 2nd edn, Vol. One, 49–65 (Springer, 2001)
Pierce, E. et al. The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ. Microbiol. 10, 2550–2573 (2008)
Ferry, J. G. & House, C. H. The stepwise evolution of early life driven by energy conservation. Mol. Biol. Evol. 23, 1286–1292 (2006)
te Poele, E. M., Bolhuis, H. & Dijkhuizen, L. Actinomycete integrative and conjugative elements. Antonie Van Leeuwenhoek 94, 127–143 (2008)
Brochier-Armanet, C., Boussau, B., Gribaldo, S. & Forterre, P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nature Rev. Microbiol. 6, 245–252 (2008)
Waters, E. et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl Acad. Sci. USA 100, 12984–12988 (2003)
Zhaxybayeva, O. et al. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc. Natl Acad. Sci. USA 106, 5865–5870 (2009)
Dagan, T., Artzy-Randrup, Y. & Martin, W. Modular networks and cumulative impact of lateral gene transfer in prokaryote genome evolution. Proc. Natl Acad. Sci. USA 105, 10039–10044 (2008)
Teeling, H. & Glöckner, F. O. RibAlign: a software tool and database for eubacterial phylogeny based on concatenated ribosomal protein subunits. BMC Bioinform. 7, 1–6 (2006)
Huber, H. et al. Ignicoccus gen. nov., a novel genus of hyperthermophilic, chemolithoautotrophic Archaea, represented by two new species, Ignicoccus islandicus sp. nov. and Ignicoccus pacificus sp. nov. Int. J. Syst. Evol. Microbiol. 50, 2093–2100 (2000)
Ramos-Vera, W. H., Berg, I. A. & Fuchs, G. Autotrophic carbon dioxide assimilation in Thermoproteales revisited. J. Bacteriol. 191, 4286–4297 (2009)
Teufel, R., Kung, J. W., Kockelkorn, D., Alber, B. E. & Fuchs, G. 3-Hydroxypropionyl-coenzyme A dehydratase and acryloyl-coenzyme A reductase, enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in Sulfolobales. J. Bacteriol. 191, 4572–4581 (2009)
Tabor, S. & Richardson, C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl Acad. Sci. USA 82, 1074–1078 (1985)
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005)
Dawson, R. M. C., Elliott, D. C., Elliott, W. H. & Jones, K. M. Data for Biochemical Research 3rd edn (Oxford Univ. Press, 1986)
Ellis, K. J. & Morrison, J. F. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 87, 405–426 (1982)
Scamuffa, M. D. & Caprioli, R. M. Comparison of the mechanisms of two distinct aldolases from Escherichia coli grown on gluconeogenic substrates. Biochim. Biophys. Acta 614, 583–590 (1980)
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970)
Schopfer, P., Heyno, E., Drepper, F. & Krieger-Liszkay, A. Naphthoquinone-dependent generation of superoxide radicals by quinone reductase isolated from the plasma membrane of soybean. Plant Physiol. 147, 864–878 (2008)
Gish, W. & States, D. J. Identification of protein coding regions by database similarity search. Nature Genet. 3, 266–272 (1993)
Rashid, N. et al. A novel candidate for the true fructose-1,6-bisphosphatase in archaea. J. Biol. Chem. 277, 30649–30655 (2002)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007)
Gascuel, O. & Steel, M. Neighbor-joining revealed. Mol. Biol. Evol. 23, 1997–2000 (2006)
Fitch, M. W. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Zool. 20, 406–416 (1971)
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981)
Witke, C. & Götz, F. Cloning, sequencing, and characterization of the gene encoding the class I fructose-1,6-bisphosphate aldolase of Staphylococcus carnosus . J. Bacteriol. 175, 7495–7499 (1993)
De Montigny, C. & Sygusch, J. Functional characterization of an extreme thermophilic class II fructose-1,6-bisphosphate aldolase. Eur. J. Biochem. 241, 243–248 (1996)
Sauvé, V. & Sygusch, J. Crystallization and preliminary X-ray analysis of native and selenomethionine fructose-1,6-bisphosphate aldolase from Thermus aquaticus . Acta Crystallogr. D 57, 310–313 (2001)
Sauve, V. & Sygusch, J. Molecular cloning, expression, purification, and characterization of fructose-1,6-bisphosphate aldolase from Thermus aquaticus . Protein Expr. Purif. 21, 293–302 (2001)
White, R. H. L. Aspartate semialdehyde and a 6-deoxy-5-ketohexose 1-phosphate are the precursors to the aromatic amino acids in Methanocaldococcus jannaschii . Biochemistry 43, 7618–7627 (2004)
Schofield, L. R., Patchett, M. L. & Parker, E. J. Expression, purification, and characterization of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Pyrococcus furiosus . Protein Expr. Purif. 34, 17–27 (2004)
Babul, J. & Guixe, V. Fructose bisphosphatase from Escherichia coli. Purification and characterization. Arch. Biochem. Biophys. 225, 944–949 (1983)
Donahue, J. L., Bownas, J. L., Niehaus, W. G. & Larson, T. J. Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol 3-phosphate regulon of Escherichia coli . J. Bacteriol. 182, 5624–5627 (2000)
Fujita, Y. & Freese, E. Purification and properties of fructose-1,6-bisphosphatase of Bacillus subtilis . J. Biol. Chem. 254, 5340–5349 (1979)
Stec, B., Yang, H., Johnson, K. A., Chen, L. & Roberts, M. F. MJ0109 is an enzyme that is both an inositol monophosphatase and the ‘missing’ archaeal fructose-1,6-bisphosphatase. Nature Struct. Biol. 7, 1046–1050 (2000)
Falb, M. et al. Metabolism of halophilic archaea. Extremophiles 12, 177–196 (2008)
Erb, T. J., Retey, J., Fuchs, G. & Alber, B. E. Ethylmalonyl-CoA mutase from Rhodobacter sphaeroides defines a new subclade of coenzyme B12-dependent acyl-CoA mutases. J. Biol. Chem. 283, 32283–32293 (2008)
Acknowledgements
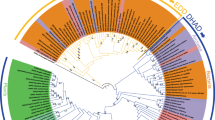
We thank R. Thauer, S. Ragsdale, H. Atomi and H. Huber for the supply of strains; F. Drepper for help with bioinformatic approaches; W. Haehnel and B. Knapp for MS analysis; N. Gad’on and C. Ebenau-Jehle for growing cells and maintaining the laboratory running; the DOE Joint Genome Institute for early release of archaeal genomic sequence data; and the Deutsche Forschungsgemeinschaft and Evonik-Degussa for financial support. We also thank M. Ziemski and I. Berg for help with data base analysis as the basis of Fig. 1a, and H. Teeling and F. O. Glöckner for providing Fig. 1b.
Author Contributions R.F.S. performed the experiments; R.F.S. and G.F. designed the work and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figure SI1-SI5 with legends, Supplementary Tables SI1-SI3 and Supplementary References. (PDF 942 kb)
Rights and permissions
About this article
Cite this article
Say, R., Fuchs, G. Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature 464, 1077–1081 (2010). https://doi.org/10.1038/nature08884
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08884
This article is cited by
-
Culexarchaeia, a novel archaeal class of anaerobic generalists inhabiting geothermal environments
ISME Communications (2022)
-
Genomic and transcriptomic dissection of Theionarchaea in marine ecosystem
Science China Life Sciences (2022)
-
The aldolase inhibitor aldometanib mimics glucose starvation to activate lysosomal AMPK
Nature Metabolism (2022)
-
Undinarchaeota illuminate DPANN phylogeny and the impact of gene transfer on archaeal evolution
Nature Communications (2020)
-
The Emergence of Life
Space Science Reviews (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.