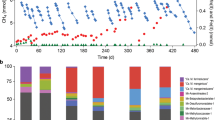
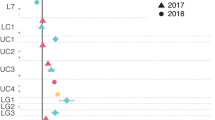
Abstract
Only three biological pathways are known to produce oxygen: photosynthesis, chlorate respiration and the detoxification of reactive oxygen species. Here we present evidence for a fourth pathway, possibly of considerable geochemical and evolutionary importance. The pathway was discovered after metagenomic sequencing of an enrichment culture that couples anaerobic oxidation of methane with the reduction of nitrite to dinitrogen. The complete genome of the dominant bacterium, named ‘Candidatus Methylomirabilis oxyfera’, was assembled. This apparently anaerobic, denitrifying bacterium encoded, transcribed and expressed the well-established aerobic pathway for methane oxidation, whereas it lacked known genes for dinitrogen production. Subsequent isotopic labelling indicated that ‘M. oxyfera’ bypassed the denitrification intermediate nitrous oxide by the conversion of two nitric oxide molecules to dinitrogen and oxygen, which was used to oxidize methane. These results extend our understanding of hydrocarbon degradation under anoxic conditions and explain the biochemical mechanism of a poorly understood freshwater methane sink. Because nitrogen oxides were already present on early Earth, our finding opens up the possibility that oxygen was available to microbial metabolism before the evolution of oxygenic photosynthesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
Sequencing and proteomic data are deposited at the National Centre for Biotechnology Information under accession numbers FP565575, SRR023516.1, SRR022749.2, GSE18535, SRR022748.2, PSE127 and PSE128.
References
Galloway, J. N. et al. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892 (2008)
Bodelier, P. L. E. & Laanbroek, H. J. Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol. Ecol. 47, 265–277 (2004)
Raghoebarsing, A. A. et al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440, 918–921 (2006)
Ettwig, K. F., van Alen, T., van de Pas-Schoonen, K. T., Jetten, M. S. M. & Strous, M. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75, 3656–3662 (2009)
Hu, S. et al. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Envir. Microbiol. Rep. 1, 377–384 (2009)
Rappé, M. S. & Giovannoni, S. J. The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394 (2003)
Thauer, R. K. & Shima, S. Methane as fuel for anaerobic microorganisms. Ann. NY Acad. Sci. 1125, 158–170 (2008)
Trotsenko, Y. A. & Murrell, J. C. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63, 183–229 (2008)
Krüger, M. et al. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426, 878–881 (2003)
Knittel, K. & Boetius, A. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334 (2009)
Ettwig, K. F. et al. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea . Environ. Microbiol. 10, 3164–3173 (2008)
Wilmes, P., Simmons, S. L., Denef, V. J. & Banfield, J. F. The dynamic genetic repertoire of microbial communities. FEMS Microbiol. Rev. 33, 109–132 (2009)
Farrer, R. A., Kemen, E., Jones, J. D. G. & Studholme, D. J. De novo assembly of the Pseudomonas syringae pv. syringae B728a genome using Illumina/Solexa short sequence reads. FEMS Microbiol. Lett. 291, 103–111 (2009)
Dutilh, B. E., Huynen, M. A. & Strous, M. Increasing the coverage of a metapopulation consensus genome by iterative read mapping and assembly. Bioinformatics 25, 2878–2881 (2009)
Zumft, W. G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 (1997)
Heider, J. Adding handles to unhandy substrates: anaerobic hydrocarbon activation mechanisms. Curr. Opin. Chem. Biol. 11, 188–194 (2007)
Chistoserdova, L., Kalyuzhnaya, M. G. & Lidstrom, M. E. C1-transfer modules: from genomics to ecology. ASM News 71, 521–528 (2005)
Howard, C. S. & Daniels, F. Stability of nitric oxide over a long time interval. J. Phys. Chem. 62, 360–361 (1958)
Parvulescu, V. I., Grange, P. & Delmon, B. Catalytic removal of NO. Catal. Today 46, 233–316 (1998)
van Ginkel, C. G., Rikken, G. B., Kroon, A. G. M. & Kengen, S. W. M. Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch. Microbiol. 166, 321–326 (1996)
Bab-Dinitz, E., Shmuely, H., Maupin-Furlow, J., Eichler, J. & Shaanan, B. Haloferax volcanii PitA: an example of functional interaction between the Pfam chlorite dismutase and antibiotic biosynthesis monooxygenase families? Bioinformatics 22, 671–675 (2006)
Zumft, W. G. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme–copper oxidase type. J. Inorg. Biochem. 99, 194–215 (2005)
Zumft, W. G. & Kroneck, P. M. H. in Advances in Microbial Physiology Vol. 52 (ed. Poole, R. K.) 107–227 (Academic, 2007)
Prior, S. D. & Dalton, H. Acetylene as a suicide substrate and active-site probe for methane monooxygenase from Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 29, 105–109 (1985)
Kool, D. M., Wrage, N., Oenema, O., Dolfing, J. & Van Groenigen, J. W. Oxygen exchange between (de)nitrification intermediates and H2O and its implications for source determination of NO3 - and N2O: a review. Rapid Commun. Mass Spectrom. 21, 3569–3578 (2007)
Demicheli, V., Quijano, C., Alvarez, B. & Radi, R. Inactivation and nitration of human superoxide dismutase (SOD) by fluxes of nitric oxide and superoxide. Free Radic. Biol. Med. 42, 1359–1368 (2007)
Allen, M. B. & van Niel, C. B. Experiments on bacterial denitrification. J. Bacteriol. 64, 397–412 (1952)
Broda, E. The history of inorganic nitrogen in the biosphere. J. Mol. Evol. 7, 87–100 (1975)
Fenchel, T. Origin and Early Evolution of Life (Oxford Univ. Press, 2002)
Castresana, J. in Respiration in Archaea and Bacteria: Diversity of Prokaryotic Electron Transport Carriers Vol. 15 (ed. Zannoni, D.) 1–14 (Springer, 2004)
Ducluzeau, A. L. et al. Was nitric oxide the first deep electron sink? Trends Biochem. Sci. 34, 9–15 (2009)
Chapman, D. J. & Schopf, J. W. Biological and Biochemical Effects of the Development of an Aerobic Environment (Princeton Univ. Press, 1983)
Holland, H. D. The oxygenation of the atmosphere and oceans. Phil. Trans. R. Soc. B 361, 903–915 (2006)
Pavlov, A. A., Kasting, J. F., Brown, L. L., Rages, K. A. & Freedman, R. Greenhouse warming by CH4 in the atmosphere of early Earth. J. Geophys. Res. Planets 105, 11981–11990 (2000)
Hayes, J. M. in Earth’s Earliest Biosphere (ed. Schopf, J. W.) 291–301 (Princeton Univ. Press, 1983)
Zhou, J., Bruns, M. A. & Tiedje, J. M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62, 316–322 (1996)
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005)
Vallenet, D. et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 34, 53–65 (2006)
Wilm, M. et al. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379, 466–469 (1996)
Andersen, K., Kjær, T. & Revsbech, N. P. An oxygen insensitive microsensor for nitrous oxide. Sens. Actuators B Chem. 81, 42–48 (2001)
Revsbech, N. P. An oxygen microsensor with a guard cathode. Limnol. Oceanogr. 34, 474–478 (1989)
Schreiber, F., Polerecky, L. & de Beer, D. Nitric oxide microsensor for high spatial resolution measurements in biofilms and sediments. Anal. Chem. 80, 1152–1158 (2008)
Gordon, D., Abajian, C. & Green, P. Consed: a graphical tool for sequence finishing. Genome Res. 8, 195–202 (1998)
Hakemian, A. S. & Rosenzweig, A. C. The biochemistry of methane oxidation. Annu. Rev. Biochem. 76, 223–241 (2007)
Stoecker, K. et al. Cohn’s Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc. Natl Acad. Sci. USA 103, 2363–2367 (2006)
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003)
Ishihama, Y., Rappsilber, J., Andersen, J. S. & Mann, M. Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 (2002)
Weatherly, D. B. et al. A heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol. Cell. Proteomics 4, 762–772 (2005)
Raghoebarsing, A. A. New Directions in Microbial Methane Oxidation. PhD thesis, Radboud Univ. Nijmegen (2006)
Friedman, S. H., Massefski, W. & Hollocher, T. C. Catalysis of intermolecular oxygen atom transfer by nitrite dehydrogenase of Nitrobacter agilis . J. Biol. Chem. 261, 538–543 (1986)
McIlvin, M. R. & Altabet, M. A. Chemical conversion of nitrate and nitrite to nitrous oxide for nitrogen and oxygen isotopic analysis in freshwater and seawater. Anal. Chem. 77, 5589–5595 (2005)
Acknowledgements
We thank F. Stams and N. Tan for sharing their ideas on NO decomposition; D. Speth and L. Russ for pilot experiments; N. Kip for providing M. acidophilus cultures; A. Pierik for electron paramagnetic resonance analysis; G. Klockgether and G. Lavik for technical assistance; and B. Kartal, J. Keltjens, A. Pol, J. van de Vossenberg and F. Widdel for helpful discussions. M.M.M.K., F.S., J.Z. and D.d.B. were supported by the Max Planck Society, M.S.M.J. by European Research Council grant 232937, M.S., K.F.E. and M.K.B. by a Vidi grant to M.S. from the Netherlands Organisation for Scientific Research (NWO), and M.L.W. and B.D. by a Horizon grant (050-71-058) from NWO.
Author Contributions Genome sequencing and assembly from enrichment culture ‘Twente’ was performed by D.L.P., E.P., S.M. and J.W. M.S., E.M.J.-M., K.-J.F. and H.S. performed the sequencing and initial assembly of sequence from enrichment culture ‘Ooij’. Mapping of sequences from enrichment culture ‘Ooij’ to ‘Twente’ was performed by B.E.D. and M.S. B.E.D. and E.P. performed SNP and coverage analyses. Genome annotation and phylogenetic analysis were conducted by M.K.B. H.J.M.O.d.C. provided support with alignments. Sample preparation for proteome analysis was performed by M.K.B. and M.W., with LC–MS/MS and protein identification performed by J.G. and H.J.C.T.W. Material for transcriptome analysis was prepared by T.v.A. and F.L., with sequencing performed by E.M.J.-M., K.-J.F. and H.S. Continuous cultures were set up and maintained by K.F.E. and K.T.v.d.P.-S. Experiments for nitrogenous intermediates were designed and performed by K.F.E., M.M.M.K., F.S., D.d.B. and J.Z., and those for methane activation were designed and performed by K.F.E. Pilot experiments were conducted by K.F.E., F.L., M.K.B., K.T.v.d.P.-S, T.A. and M.S. K.F.E., M.K.B., M.S.M.J. and M.S. conceived the research. K.F.E., M.K.B. and M.S. wrote the paper with input from all other authors.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-5 and Supplementary Figures 1-7 with legends. (PDF 530 kb)
Rights and permissions
About this article
Cite this article
Ettwig, K., Butler, M., Le Paslier, D. et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548 (2010). https://doi.org/10.1038/nature08883
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08883
This article is cited by
-
Methane-dependent complete denitrification by a single Methylomirabilis bacterium
Nature Microbiology (2024)
-
Vertical distribution of Candidatus Methylomirabilis and Methanoperedens in agricultural soils
Applied Microbiology and Biotechnology (2024)
-
Effect of no-till followed by crop diversification on the soil microbiome in a boreal short cereal rotation
Biology and Fertility of Soils (2024)
-
Stratified microbial communities in Australia’s only anchialine cave are taxonomically novel and drive chemotrophic energy production via coupled nitrogen-sulphur cycling
Microbiome (2023)
-
Phylogenetic and metabolic diversity of microbial communities performing anaerobic ammonium and methane oxidations under different nitrogen loadings
ISME Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.