Abstract
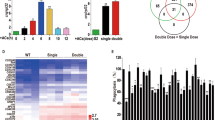
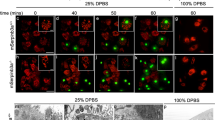
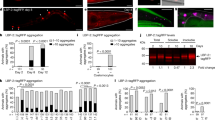
Engulfment of apoptotic cells occurs throughout life in multicellular organisms. Impaired apoptotic cell clearance (due to defective recognition, internalization or degradation) results in autoimmune disease1,2. One fundamental challenge in understanding how defects in corpse removal translate into diseased states is the identification of critical components orchestrating the different stages of engulfment. Here we use genetic, cell biological and molecular studies in Caenorhabditis elegans and mammalian cells to identify SAND-1 and its partner CCZ-1 as new factors in corpse removal. In worms deficient in either sand-1 or ccz-1, apoptotic cells are internalized and the phagosomes recruit the small GTPase RAB-5 but fail to progress to the subsequent RAB-7(+) stage. The mammalian orthologues of SAND-1, namely Mon1a and Mon1b, were similarly required for phagosome maturation. Mechanistically, Mon1 interacts with GTP-bound Rab5, identifying Mon1 as a previously unrecognized Rab5 effector. Moreover, a Mon1–Ccz1 complex (but not either protein alone) could bind Rab7 and could also influence Rab7 activation, suggesting Mon1–Ccz1 as an important link in progression from the Rab5-positive stage to the Rab7-positive stage of phagosome maturation. Taken together, these data identify SAND-1 (Mon1) and CCZ-1 (Ccz1) as critical and evolutionarily conserved components regulating the processing of ingested apoptotic cell corpses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Albert, M. L. Death-defying immunity: do apoptotic cells influence antigen processing and presentation? Nature Rev. Immunol. 4, 223–231 (2004)
Ravichandran, K. S. & Lorenz, U. Engulfment of apoptotic cells: signals for a good meal. Nature Rev. Immunol. 7, 964–974 (2007)
Lettre, G. & Hengartner, M. O. Developmental apoptosis in C. elegans: a complex CEDnario. Nature Rev. Mol. Cell Biol. 7, 97–108 (2006)
Gumienny, T. L., Lambie, E., Hartwieg, E., Horvitz, H. R. & Hengartner, M. O. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126, 1011–1022 (1999)
Kinchen, J. M. & Hengartner, M. O. Tales of cannibalism, suicide, and murder: Programmed cell death in C. elegans . Curr. Top. Dev. Biol. 65, 1–45 (2005)
Mangahas, P. M. & Zhou, Z. Clearance of apoptotic cells in Caenorhabditis elegans . Semin. Cell Dev. Biol. 16, 295–306 (2005)
Kinchen, J. M. et al. A pathway for phagosome maturation during engulfment of apoptotic cells. Nature Cell Biol. 10, 556–566 (2008)
Gengyo-Ando, K. et al. The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans . EMBO Rep. 8, 152–157 (2007)
Grosshans, B. L., Ortiz, D. & Novick, P. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl Acad. Sci. USA 103, 11821–11827 (2006)
Poteryaev, D., Fares, H., Bowerman, B. & Spang, A. Caenorhabditis elegans SAND-1 is essential for RAB-7 function in endosomal traffic. EMBO J. 26, 301–312 (2007)
Hoeppner, D. J. et al. eor-1 and eor-2 are required for cell-specific apoptotic death in C. elegans . Dev. Biol. 274, 125–138 (2004)
Zhou, Z., Hartwieg, E. & Horvitz, H. R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans . Cell 104, 43–56 (2001)
Huynh, K. K. et al. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 26, 313–324 (2007)
Kinchen, J. M. & Ravichandran, K. S. Phagosome maturation: going through the acid test. Nature Rev. Mol. Cell Biol. 9, 781–795 (2008)
Hackam, D. J. et al. Regulation of phagosomal acidification. Differential targeting of Na+/H+ exchangers, Na+/K+-ATPases, and vacuolar-type H+-ATPases. J. Biol. Chem. 272, 29810–29820 (1997)
Lettre, G. et al. Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C. elegans . Cell Death Differ. 11, 1198–1203 (2004)
Yu, X., Lu, N. & Zhou, Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 6, e61 (2008)
Yu, X., Odera, S., Chuang, C. H., Lu, N. & Zhou, Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev. Cell 10, 743–757 (2006)
Lu, Q. et al. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development 135, 1069–1080 (2008)
Rink, J., Ghigo, E., Kalaidzidis, Y. & Zerial, M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749 (2005)
Li, W. et al. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development 136, 2445–2455 (2009)
Wang, C. W., Stromhaug, P. E., Kauffman, E. J., Weisman, L. S. & Klionsky, D. J. Yeast homotypic vacuole fusion requires the Ccz1–Mon1 complex during the tethering/docking stage. J. Cell Biol. 163, 973–985 (2003)
Sun, J. et al. Mycobacterium bovis BCG disrupts the interaction of Rab7 with RILP contributing to inhibition of phagosome maturation. J. Leukoc. Biol. 82, 1437–1445 (2007)
D’Adamo, P. et al. Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nature Genet. 19, 134–139 (1998)
Jekely, G., Sung, H. H., Luque, C. M. & Rorth, P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev. Cell 9, 197–207 (2005)
Mosesson, Y., Mills, G. B. & Yarden, Y. Derailed endocytosis: an emerging feature of cancer. Nature Rev. Cancer 8, 835–850 (2008)
Rock, K. L. & Shen, L. Cross-presentation: underlying mechanisms and role in immune surveillance. Immunol. Rev. 207, 166–183 (2005)
Gruenberg, J. & van der Goot, F. G. Mechanisms of pathogen entry through the endosomal compartments. Nature Rev. Mol. Cell Biol. 7, 495–504 (2006)
Kinchen, J. M. et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans . Nature 434, 93–99 (2005)
Grimsley, C. M., Lu, M., Haney, L. B., Kinchen, J. M. & Ravichandran, K. S. Characterization of a novel interaction between ELMO1 and ERM proteins. J. Biol. Chem. 281, 5928–5937 (2006)
Brenner, S. The genetics of Caenorhabditis elegans . Genetics 77, 71–94 (1974)
Feng, Y., Press, B. & Wandinger-Ness, A. Rab 7: an important regulator of late endocytic membrane traffic. J. Cell Biol. 131, 1435–1452 (1995)
Kamath, R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003)
Tosello-Trampont, A. C. et al. Identification of two signaling submodules within the CrkII/ELMO/Dock180 pathway regulating engulfment of apoptotic cells. Cell Death Differ. 14, 963–972 (2007)
Tosello-Trampont, A. C., Brugnera, E. & Ravichandran, K. S. Evidence for a conserved role for CRKII and Rac in engulfment of apoptotic cells. J. Biol. Chem. 276, 13797–13802 (2001)
Acknowledgements
We thank J. Casanova, C. Grimsley and members of the Ravichandran laboratory for helpful conversations; the Caenorhabditis Genetics Consortium (CGC) for nematode strains; A. Wandinger-Ness for Rab7 expression constructs; and J. Redick and S. Guillot of the Advanced Microscopy Facility for the preparation of specimens for electron microscopy. This work was supported by a post-doctoral fellowship via an NIH T32 Immunology Training Grant and American Heart Association Award (to J.M.K.), and grants from the NIGMS/NIH (to K.S.R.). K.S.R. is a William Benter Senior Fellow of the American Asthma Foundation.
Author Contributions J.M.K. performed all the experiments. J.M.K and K.S.R. planned and analysed the experimental results and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1-S12 with legends and Supplementary Table S1. (PDF 7990 kb)
Rights and permissions
About this article
Cite this article
Kinchen, J., Ravichandran, K. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 464, 778–782 (2010). https://doi.org/10.1038/nature08853
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08853
This article is cited by
-
An account of conserved functions and how biologists use them to integrate cell and evolutionary biology
Biology & Philosophy (2023)
-
The PripA-TbcrA complex-centered Rab GAP cascade facilitates macropinosome maturation in Dictyostelium
Nature Communications (2022)
-
SUMO modification in apoptosis
Journal of Molecular Histology (2021)
-
A trimeric Rab7 GEF controls NPC1-dependent lysosomal cholesterol export
Nature Communications (2020)
-
EFF-1 fusogen promotes phagosome sealing during cell process clearance in Caenorhabditis elegans
Nature Cell Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.