Abstract
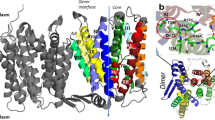
In extremely acidic environments, enteric bacteria such as Escherichia coli rely on the amino acid antiporter AdiC to expel protons by exchanging intracellular agmatine (Agm2+) for extracellular arginine (Arg+)1,2,3. AdiC is a representative member of the amino acid-polyamine-organocation (APC) superfamily of membrane transporters4,5. The structure of substrate-free AdiC revealed a homodimeric assembly, with each protomer containing 12 transmembrane segments and existing in an outward-open conformation6,7. The overall folding of AdiC is similar to that of the Na+-coupled symporters8,9,10,11. Despite these advances, it remains unclear how the substrate (arginine or agmatine) is recognized and transported by AdiC. Here we report the crystal structure of an E. coli AdiC variant bound to Arg at 3.0 Å resolution. The positively charged Arg is enclosed in an acidic binding chamber, with the head groups of Arg hydrogen-bonded to main chain atoms of AdiC and the aliphatic portion of Arg stacked by hydrophobic side chains of highly conserved residues. Arg binding induces pronounced structural rearrangement in transmembrane helix 6 (TM6) and, to a lesser extent, TM2 and TM10, resulting in an occluded conformation. Structural analysis identified three potential gates, involving four aromatic residues and Glu 208, which may work in concert to differentially regulate the upload and release of Arg and Agm.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Foster, J. W. Escherichia coli acid resistance: tales of an amateur acidophile. Nature Rev. Microbiol. 2, 898–907 (2004)
Iyer, R., Williams, C. & Miller, C. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli . J. Bacteriol. 185, 6556–6561 (2003)
Gong, S., Richard, H. & Foster, J. W. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli . J. Bacteriol. 185, 4402–4409 (2003)
Jack, D. L., Paulsen, I. T. & Saier, M. H. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146, 1797–1814 (2000)
Casagrande, F. et al. Projection structure of a member of the amino acid/polyamine/organocation transporter superfamily. J. Biol. Chem. 283, 33240–33248 (2008)
Gao, X. et al. Structure and mechanism of an amino acid antiporter. Science 324, 1565–1568 (2009)
Fang, Y. et al. Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature 460, 1040–1043 (2009)
Yamashita, A., Singh, S. K., Kawate, T., Jin, Y. & Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl–-dependent neurotransmitter transporters. Nature 437, 215–223 (2005)
Ressl, S., Terwisscha van Scheltinga, A. C., Vonrhein, C., Ott, V. & Ziegler, C. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature 458, 47–52 (2009)
Faham, S. et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814 (2008)
Weyand, S. et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322, 709–713 (2008)
Hersh, B. M., Farooq, F. T., Barstad, D. N., Blankenhorn, D. L. & Slonczewski, J. L. A glutamate-dependent acid resistance gene in Escherichia coli . J. Bacteriol. 178, 3978–3981 (1996)
Castanie-Cornet, M. P., Penfound, T. A., Smith, D., Elliott, J. F. & Foster, J. W. Control of acid resistance in Escherichia coli . J. Bacteriol. 181, 3525–3535 (1999)
Soksawatmaekhin, W., Kuraishi, A., Sakata, K., Kashiwagi, K. & Igarashi, K. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli . Mol. Microbiol. 51, 1401–1412 (2004)
Soksawatmaekhin, W., Uemura, T., Fukiwake, N., Kashiwagi, K. & Igarashi, K. Identification of the cadaverine recognition site on the cadaverine-lysine antiporter CadB. J. Biol. Chem. 281, 29213–29220 (2006)
Kashiwagi, K. et al. Identification of the putrescine recognition site on polyamine transport protein PotE. J. Biol. Chem. 275, 36007–36012 (2000)
Kashiwagi, K., Miyamoto, S., Suzuki, F., Kobayashi, H. & Igarashi, K. Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli . Proc. Natl Acad. Sci. USA 89, 4529–4533 (1992)
Kashiwagi, K., Shibuya, S., Tomitori, H., Kuraishi, A. & Igarashi, K. Excretion and uptake of putrescine by the PotE protein in Escherichia coli . J. Biol. Chem. 272, 6318–6323 (1997)
Shaffer, P. L., Goehring, A., Shankaranarayanan, A. & Gouaux, E. Structure and mechanism of a Na+-independent amino acid transporter. Science 325, 1010–1014 (2009)
Fang, Y., Kolmakova-Partensky, L. & Miller, C. A bacterial arginine-agmatine exchange transporter involved in extreme acid resistance. J. Biol. Chem. 282, 176–182 (2007)
Singh, S. K., Piscitelli, C. L., Yamashita, A. & Gouaux, E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science 322, 1655–1661 (2008)
Zhou, Z. et al. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science 317, 1390–1393 (2007)
Singh, S. K., Yamashita, A. & Gouaux, E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature 448, 952–956 (2007)
Forrest, L. R. et al. Mechanism for alternating access in neurotransmitter transporters. Proc. Natl Acad. Sci. USA 105, 10338–10343 (2008)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
DeLano, W. L. The PyMOL Molecular Graphics System. 〈http://www.pymol.org〉 (2002)
Cowtan, K. dm: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsl. Protein Crystallogr. 31, 34–38 (1994)
Acknowledgements
We thank C. Miller and Y. Xiong for discussion and exchange of information, N. Shimizu, T. Kumasaka and S. Baba at the Spring-8 beamline BL41XU for on-site assistance, and N. Yan for discussion and comments on the manuscript. This work was supported by the Ministry of Science and Technology (grant no. 2009CB918801), Tsinghua University 985 Phase II funds, National Natural Science Foundation, and Beijing Municipal Commissions of Education and Science and Technology.
Author Contributions Experiments were performed by X.G., L.Z., X.J., F.L., C.Y., X.Z. and J.W. Data were analysed by X.G., L.Z., X.J., J.W. and Y.S. The manuscript was prepared by X.G., L.Z. and Y.S.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-3, Supplementary Figures 1-12 with Legends and Supplementary References. (PDF 2651 kb)
Rights and permissions
About this article
Cite this article
Gao, X., Zhou, L., Jiao, X. et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 463, 828–832 (2010). https://doi.org/10.1038/nature08741
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08741
This article is cited by
-
Cryo-EM structure of the human Asc-1 transporter complex
Nature Communications (2024)
-
Conformational transition induced in the aspartate:alanine antiporter by l-Ala binding
Scientific Reports (2022)
-
Heteromeric Amino Acid Transporters in Brain: from Physiology to Pathology
Neurochemical Research (2022)
-
High-resolution structure of the amino acid transporter AdiC reveals insights into the role of water molecules and networks in oligomerization and substrate binding
BMC Biology (2021)
-
Asc-1 Transporter (SLC7A10): Homology Models And Molecular Dynamics Insights Into The First Steps Of The Transport Mechanism
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.