Abstract
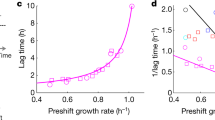
An important challenge in systems biology is to quantitatively describe microbial growth using a few measurable parameters that capture the essence of this complex phenomenon. Two key events at the cell membrane—extracellular glucose sensing and uptake—initiate the budding yeast’s growth on glucose. However, conventional growth models focus almost exclusively on glucose uptake. Here we present results from growth-rate experiments that cannot be explained by focusing on glucose uptake alone. By imposing a glucose uptake rate independent of the sensed extracellular glucose level, we show that despite increasing both the sensed glucose concentration and uptake rate, the cell’s growth rate can decrease or even approach zero. We resolve this puzzle by showing that the interaction between glucose perception and import, not their individual actions, determines the central features of growth, and characterize this interaction using a quantitative model. Disrupting this interaction by knocking out two key glucose sensors significantly changes the cell’s growth rate, yet uptake rates are unchanged. This is due to a decrease in burden that glucose perception places on the cells. Our work shows that glucose perception and import are separate and pivotal modules of yeast growth, the interaction of which can be precisely tuned and measured.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Monod, J. Recherches sur la Croissance des Cultures Bacteriennes (Hermann et Cie, 1942)
Bennett, M. R. et al. Metabolic gene regulation in a dynamically changing environment. Nature 454, 1119–1122 (2008)
Zaslaver, A. et al. Just-in-time transcription program in metabolic pathways. Nature Genet. 36, 486–491 (2004)
Airoldi, E. et al. Predicting cellular growth from gene expression signatures. PLoS Comp. Biol. 5, e1000257 (2009)
Dekel, E. & Alon, U. Optimality and evolutionary tuning of the expression level of a protein. Nature 436, 588–592 (2005)
Krishna, S., Semssey, S. & Sneppen, K. Combinatorics of feedback in cellular uptake and metabolism of small molecules. Proc. Natl Acad. Sci. USA 104, 20815–20819 (2007)
Ihmels, J., Levy, R. & Barkai, N. Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae . Nature Biotechnol. 22, 86–92 (2003)
Famili, I., Forster, J., Nielsen, J. & Palsson, B. O. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc. Natl Acad. Sci. USA 100, 13134–13139 (2003)
Bilu, Y., Shlomi, T., Barkai, N. & Ruppin, E. Conservation of expression and sequence of metabolic genes is reflected by activity across metabolic states. PLoS Comp. Biol. 2, e106.
Levine, E. & Hwa, T. Stochastic fluctuations in metabolic pathways. Proc. Natl Acad. Sci. USA 104, 9224–9229 (2007)
Fell, D. A. Understanding the Control of Metabolism (Portland, 1997)
Savageau, M. A. Biochemical Systems Analysis: A Study of Function and Design in Molecular Biology (Addison-Wesely, 1976)
Goyal, S. & Wingreen, N. S. Growth-induced instability in metabolic networks. Phys. Rev. Lett. 98, 138105 (2007)
Nielsen, J., Villadsen, J. & Liden, G. Bioreaction Engineering Principles 235–311 (Springer, 2003)
Dickinson, J. R. & Schweizer, M. The Metabolism and Molecular Physiology of Saccharomyces cerevisiae (CRC, 2004)
Alon, U. Simplicity in biology. Nature 446, 497 (2007)
Mallavarapu, A., Thomson, M., Ullian, B. & Gunawardena, J. Programming with models: modularity and abstraction provide powerful capabilities for systems biology. J. R. Soc. Interface 6, 257–270 (2009)
Moriya, H. & Johnston, M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl Acad. Sci. USA 101, 1572–1577 (2004)
Boles, E. & Hollenberg, C. P. The molecular genetics of hexose transport in yeasts. FEMS Microbiol. Rev. 21, 85–111 (1997)
Reifenberger, E., Freidel, K. & Ciriacy, M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol. Microbiol. 16, 157–167 (1995)
Bisson, L. F., Coons, D. M., Kruckeberg, A. L. & Lewis, D. A. Yeast sugar transporters. Crit. Rev. Biochem. Mol. Biol. 28, 259–308 (1993)
Ozcan, S. & Johnston, M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Microbiol. Mol. Biol. Rev. 63, 554–569 (1999)
Pao, S. S., Paulsen, I. T. & Saier, M. H. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34 (1998)
Wieczorke, R. et al. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae . FEBS Lett. 464, 123–128 (1999)
Reifenberger, E., Boles, E. & Ciriacy, M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Eur. J. Biochem. 245, 324–333 (1997)
Maier, A., Volker, B., Boles, E. & Fuhrmann, G. F. Characterisation of glucose transport in Saccharomyces cerevisiae with plasma membrane vesicles (countertransport) and intact cells (initial uptake) with single Hxt1, Hxt2, Hxt3, Hxt4, Hxt6, Hxt7 or Gal2 transporters. FEMS Yeast Res. 2, 539–550 (2002)
Walsh, M. C., Scholte, M., Valkier, J., Smits, H. P. & van Dam, K. Glucose sensing and signaling properties in Saccharomyces cerevisiae require the presence of at least two members of the glucose transporter family. J. Bacteriol. 170, 2593–2597 (1996)
Jiang, Y., Davis, C. & Broach, J. Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 17, 6942–6951 (1998)
Boer, V. M., Amini, S. & Botstein, D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc. Natl Acad. Sci. USA 105, 6930–6935 (2008)
van Hoek, P., van Dijken, J. & Pronk, J. Effects of specific growth rate on fermentative capacity of baker’s yeast. Appl. Environ. Microbiol. 64, 4226–4233 (1998)
Reijenga, K. A. et al. Control of glycolytic dynamics by hexose transport in Saccharomyces cerevisiae . Biophys. J. 80, 626–634 (2001)
Kaniak, A., Xue, Z., Macool, D., Kim, J. H. & Johnston, M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae . Eukaryot. Cell 3, 221–231 (2004)
Gancedo, J. M. The early steps of glucose signaling in yeast. FEMS Microbiol. Rev. 32, 673–704 (2008)
Kim, J. H. & Johnston, M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevsiae . J. Biol. Chem. 281, 26144–26149 (2006)
Santangelo, G. M. Glucose signaling in Saccharomyces cerevisiae . Microbiol. Mol. Biol. Rev. 70, 253–282 (2006)
Levy, S. et al. Strategy of transcription regulation in the budding yeast. PLoS One 2, e250 (2007)
Yin, Z. et al. Glucose triggers different global responses in yeast, depending on the strength of the signal, and transiently stabilizes ribosomal protein mRNAs. Mol. Microbiol. 48, 713–724 (2003)
Stephanopoulos, G. Challenges in engineering microbes for biofuels production. Science 315, 801–804 (2007)
Lorenz, D. R., Cantor, C. R. & Collins, J. J. A network biology approach to aging in yeast. Proc. Natl Acad. Sci. USA 106, 1145–1150 (2009)
Ostergaard, S., Olsson, L. & Nielsen, J. Metabolic engineering of Saccharomyces cerevisiae . Microbiol. Mol. Biol. Rev. 64, 34–50 (2000)
Kell, D. B. Metabolomics and systems biology: making sense of the soup. Curr. Opin. Microbiol. 7, 296–307 (2004)
Savageau, M. A., Coelho, P., Fasani, R., Tolla, D. & Salvador, A. Phenotypes and tolerances in the design space of biochemical systems. Proc. Natl Acad. Sci. USA 106, 6435–6440 (2009)
Ihmels, J. et al. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309, 938–940 (2005)
Klumpp, S. & Hwa, T. Growth-rate-dependent partitioning of RNA polymerases in bacteria. Proc. Natl Acad. Sci. USA 105, 20245–20250 (2008)
Duarte, N. C., Palsson, B., Ø & Fu, P. Integrated analysis of metabolic phenotypes in Saccharomyces cerevisiae . BMC Genomics 5, 63 (2004)
Daran-Lapujade, P. et al. The fluxes through glycolytic enzymes in Saccharomyces cereivisiae are predominantly regulated at posttranscriptional levels. Proc. Natl Acad. Sci. USA 104, 15753–15758 (2007)
Castrillo, J. I. et al. Growth control of the eukaryote cell: a systems biology study in yeast. J. Biol. 6, 4 (2007)
Stelling, J. Mathematical models in microbial systems biology. Curr. Opin. Microbiol. 7, 513–518 (2004)
Sheff, M. & Thorn, K. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae . Yeast 21, 661–670 (2004)
Kim, J. H., Polish, J. & Johnston, M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol. Cell. Biol. 23, 5208–5216 (2003)
Acknowledgements
We thank E. Boles for the kind gift of strains. We also thank D. Botstein, D. Muzzey, J. Gore and S. Rifkin for critical reading of our manuscript and useful discussions. This work was funded by a National Institutes of Health (NIH) Director’s Pioneer awarded to A.v.O., and grants from the NIH and National Science Foundation (NSF). H.Y. was supported by the Natural Sciences and Engineering Research Council of Canada’s (NSERC) Graduate Fellowship.
Author Contributions H.Y. performed the experiments. H.Y. and A.v.O. designed experiments, analysed data and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Figures 1-17 with Legends, Supplementary References and a Supplementary Chart of the Strains used in this study. (PDF 4418 kb)
Rights and permissions
About this article
Cite this article
Youk, H., van Oudenaarden, A. Growth landscape formed by perception and import of glucose in yeast. Nature 462, 875–879 (2009). https://doi.org/10.1038/nature08653
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08653
This article is cited by
-
Predicting the re-distribution of antibiotic molecules caused by inter-species interactions in microbial communities
ISME Communications (2022)
-
Role of Noise-Induced Cellular Variability in Saccharomyces cerevisiae During Metabolic Adaptation: Causes, Consequences and Ramifications
Journal of the Indian Institute of Science (2020)
-
Differential scaling between G1 protein production and cell size dynamics promotes commitment to the cell division cycle in budding yeast
Nature Cell Biology (2019)
-
Low affinity uniporter carrier proteins can increase net substrate uptake rate by reducing efflux
Scientific Reports (2018)
-
Glucose repression can be alleviated by reducing glucose phosphorylation rate in Saccharomyces cerevisiae
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.