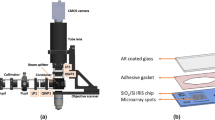
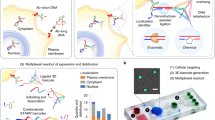
Abstract
Techniques to detect and quantify DNA and RNA molecules in biological samples have had a central role in genomics research1,2,3. Over the past decade, several techniques have been developed to improve detection performance and reduce the cost of genetic analysis4,5,6,7,8,9,10. In particular, significant advances in label-free methods have been reported11,12,13,14,15,16,17. Yet detection of DNA molecules at concentrations below the femtomolar level requires amplified detection schemes1,8. Here we report a unique nanomechanical response of hybridized DNA and RNA molecules that serves as an intrinsic molecular label. Nanomechanical measurements on a microarray surface have sufficient background signal rejection to allow direct detection and counting of hybridized molecules. The digital response of the sensor provides a large dynamic range that is critical for gene expression profiling. We have measured differential expressions of microRNAs in tumour samples; such measurements have been shown to help discriminate between the tissue origins of metastatic tumours18. Two hundred picograms of total RNA is found to be sufficient for this analysis. In addition, the limit of detection in pure samples is found to be one attomolar. These results suggest that nanomechanical read-out of microarrays promises attomolar-level sensitivity and large dynamic range for the analysis of gene expression, while eliminating biochemical manipulations, amplification and labelling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Higuchi, R., Dollinger, G., Walsh, P. S. & Griffith, R. Simultaneous amplification and detection of specific DNA sequences. Nature Biotechnol. 10, 413–417 (1992)
Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 (1995)
Lockhart, D. J. & Winzeler, E. A. Genomics, gene expression and DNA arrays. Nature 405, 827–836 (2000)
Taton, T. A., Mirkin, C. A. & Letsinger, R. L. Scanometric DNA array detection with nanoparticle probes. Science 289, 1757–1760 (2000)
Fritz, J., Cooper, E. B., Gaudet, S., Sorger, P. K. & Manalis, S. R. Electronic detection of DNA by its intrinsic molecular charge. Proc. Natl Acad. Sci. USA 99, 14142–14146 (2002)
Cao, Y. C., Jin, R. & Mirkin, C. A. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science 297, 1536–1540 (2002)
Park, S. J., Taton, T. A. & Mirkin, C. A. Array-based electrical detection of DNA with nanoparticle probes. Science 295, 1503–1506 (2002)
Nam, J. M., Stoeva, S. I. & Mirkin, C. A. Bio-bar-code-based DNA detection with PCR-like sensitivity. J. Am. Chem. Soc. 126, 5932–5933 (2004)
Fang, S., Lee, H. J., Wark, A. W. & Corn, R. M. Attomole microarray detection of microRNAs by nanoparticle-amplified SPR imaging measurements of surface polyadenylation reactions. J. Am. Chem. Soc. 128, 14044–14046 (2006)
Geiss, G. K. et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature Biotechnol. 26, 317–325 (2008)
Hahm, J.-i. & Lieber, C. M. Direct ultrasensitive electrical detection of DNA and DNA sequence variations using nanowire nanosensors. Nano Lett. 4, 51–54 (2004)
Fritz, J. et al. Translating biomolecular recognition into nanomechanics. Science 288, 316–318 (2000)
Ilic, B. et al. Enumeration of DNA molecules bound to a nanomechanical oscillator. Nano Lett. 5, 925–929 (2005)
Zhang, J. et al. Rapid and label-free nanomechanical detection of biomarker transcripts in human RNA. Nature Nanotechnol. 1, 214–220 (2006)
Mertens, J. et al. Label-free detection of DNA hybridization based on hydration-induced tension in nucleic acid films. Nature Nanotechnol. 3, 301–307 (2008)
Clack, N. G., Salaita, K. & Groves, J. T. Electrostatic readout of DNA microarrays with charged microspheres. Nature Biotechnol. 26, 825–830 (2008)
Burg, T. P. et al. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 446, 1066–1069 (2007)
Rosenfeld, N. et al. MicroRNAs accurately identify cancer tissue origin. Nature Biotechnol. 26, 462–469 (2008)
Cheng, M. M. C. et al. Nanotechnologies for biomolecular detection and medical diagnostics. Curr. Opin. Chem. Biol. 10, 11–19 (2006)
Sinensky, A. K. & Belcher, A. M. Label-free and high-resolution protein/DNA nanoarray analysis using Kelvin probe force microscopy. Nature Nanotechnol. 2, 653–659 (2007)
Ke, Y., Lindsay, S., Chang, Y., Liu, Y. & Yan, H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science 319, 180–183 (2008)
Sahin, O., Su, C., Magonov, S., Quate, C. F. & Solgaard, O. An atomic force microscope tip designed to measure time-varying nanomechanical forces. Nature Nanotechnol. 2, 507–514 (2007)
Sahin, O. & Erina, N. High-resolution and large dynamic range nanomechanical mapping in tapping-mode atomic force microscopy. Nanotechnology 19, 445717 (2008)
Zhou, D. J., Sinniah, K., Abell, C. & Rayment, T. Label-free detection of DNA hybridization at the nanoscale: a highly sensitive and selective approach using atomic-force microscopy. Angew. Chem. Int. Ed. 42, 4934–4937 (2003)
Mirmomtaz, E. et al. Nano Lett. 8, 4134–4139 (2008)
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 5, 522–531 (2004)
Chen, C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 (2005)
Demers, L. M. et al. Direct patterning of modified oligonucleotides on metals and insulators by dip-pen nanolithography. Science 296, 1836–1838 (2002)
Yu, A. A. et al. Supramolecular nanostamping: using DNA as movable type. Nano Lett. 5, 1061–1064 (2005)
Herne, T. M. & Tarlov, M. J. Characterization of DNA probes immobilized on gold surfaces. J. Am. Chem. Soc. 119, 8916–8920 (1997)
Sahin, O. Harnessing bifurcations in tapping-mode atomic force microscopy to calibrate time-varying tip-sample force measurements. Rev. Sci. Instrum. 78, 103707 (2007)
Acknowledgements
This research is supported by the Rowland Junior Fellows Program. H.H.J.P. was supported in part by US National Institutes of Health grant HG000205. We thank R. W. Davis for discussions.
Author Contributions S.H. performed the experiments, prepared gold substrates and developed experimental protocols; H.H.J.P. and O.S. contributed to the experiments. S.H. and H.H.J.P. designed and performed the surface chemistry, O.S. designed the cantilevers and wrote the stiffness calculation program, and H.H.J.P. and O.S. designed the biological assay. O.S. directed the research and wrote the paper; all authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
A patent application has been filed by Stanford University.
Supplementary information
Supplementary Information
This file contains Supplementary Data, Supplementary Notes and Supplementary Figures 1-4 and 6-8 with Legends. (PDF 1407 kb)
Supplementary Figure
This file contains Supplementary Figure 5 in its original high pixel density version. (JPG 4340 kb)
Rights and permissions
About this article
Cite this article
Husale, S., Persson, H. & Sahin, O. DNA nanomechanics allows direct digital detection of complementary DNA and microRNA targets. Nature 462, 1075–1078 (2009). https://doi.org/10.1038/nature08626
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08626
This article is cited by
-
Nanomechanical DNA resonators for sensing and structural analysis of DNA-ligand complexes
Nature Communications (2019)
-
Clinical utility of circulating non-coding RNAs — an update
Nature Reviews Clinical Oncology (2018)
-
Dual functional Phi29 DNA polymerase-triggered exponential rolling circle amplification for sequence-specific detection of target DNA embedded in long-stranded genomic DNA
Scientific Reports (2017)
-
Polystyrene Microspheres Coupled with Hybridization Chain Reaction for Dual-Amplified Chemiluminescence Detection of Specific DNA Sequences
Journal of Analysis and Testing (2017)
-
In Vivo Detection of miRNA Expression in Tumors Using an Activatable Nanosensor
Molecular Imaging and Biology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.