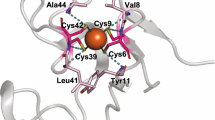
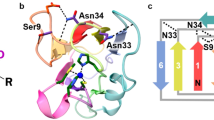
Abstract
Redox processes are at the heart of numerous functions in chemistry and biology, from long-range electron transfer in photosynthesis and respiration to catalysis in industrial and fuel cell research. These functions are accomplished in nature by only a limited number of redox-active agents. A long-standing issue in these fields is how redox potentials are fine-tuned over a broad range with little change to the redox-active site or electron-transfer properties. Resolving this issue will not only advance our fundamental understanding of the roles of long-range, non-covalent interactions in redox processes, but also allow for design of redox-active proteins having tailor-made redox potentials for applications such as artificial photosynthetic centres1,2 or fuel cell catalysts3 for energy conversion. Here we show that two important secondary coordination sphere interactions, hydrophobicity and hydrogen-bonding, are capable of tuning the reduction potential of the cupredoxin azurin over a 700 mV range, surpassing the highest and lowest reduction potentials reported for any mononuclear cupredoxin, without perturbing the metal binding site beyond what is typical for the cupredoxin family of proteins. We also demonstrate that the effects of individual structural features are additive and that redox potential tuning of azurin is now predictable across the full range of cupredoxin potentials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Beratan, D. N., Onuchic, J. N., Winkler, J. R. & Gray, H. B. Electron-tunneling pathways in proteins. Science 258, 1740–1741 (1992)
Kanan, M. W. & Nocera, D. G. In situ formation of an oxygen-evolving catalyst in neutral water containing phosphate and Co2+ . Science 321, 1072–1075 (2008)
Blanford, C. F., Heath, R. S. & Armstrong, F. A. A stable electrode for high-potential, electrocatalytic O2 reduction based on rational attachment of a blue copper oxidase to a graphite surface. Chem. Commun. 1710–1712 (2007)
Gray, H. B., Malmström, B. G. & Williams, R. J. P. Copper coordination in blue proteins. J. Biol. Inorg. Chem. 5, 551–559 (2000)
Solomon, E. I., Szilagyi, R. K., DeBeer George, S. & Basumallick, L. Electronic structures of metal sites in proteins and models: contributions to function in blue copper proteins. Chem. Rev. 104, 419–458 (2004)
Vila, A. J. & Fernández, C. O. in Handbook on Metalloproteins (eds Bertini, I., Sigel, A. & Sigel, H.) 813–856 (Marcel Dekker, 2001)
Lu, Y. in Comprehensive Coordination Chemistry II: From Biology to Nanotechnology Vol. 8, Biocoordination Chemistry (eds Que, L. & Tolman, W. B.) 91–122 (Elsevier, 2004)
Dennison, C. Investigating the structure and function of cupredoxins. Coord. Chem. Rev. 249, 3025–3054 (2005)
Solomon, E. I. Spectroscopic methods in bioinorganic chemistry: blue to green to red copper sites. Inorg. Chem. 45, 8012–8025 (2006)
Varadarajan, R., Zewert, T. E., Gray, H. B. & Boxer, S. G. Effects of buried ionizable amino acids on the reduction potential of recombinant myoglobin. Science 243, 69–72 (1989)
Battistuzzi, G., Borsari, M., Loschi, L., Righi, F. & Sola, M. Redox thermodynamics of blue copper proteins. J. Am. Chem. Soc. 121, 501–506 (1999)
Dey, A. et al. Solvent tuning of electrochemical potentials in the active sites of HiPIP versus ferredoxin. Science 318, 1464–1468 (2007)
Garner, D. K. et al. Reduction potential tuning of the blue copper center in Pseudomonas aeruginosa azurin by the axial methionine as probed by unnatural amino acids. J. Am. Chem. Soc. 128, 15608–15617 (2006)
Berry, S. M., Ralle, M., Low, D. W., Blackburn, N. J. & Lu, Y. Probing the role of axial methionine in the blue copper center of azurin with unnatural amino acids. J. Am. Chem. Soc. 125, 8760–8768 (2003)
Abdelhamid, R. F. et al. π-π interaction between aromatic ring and copper-coordinated His81 imidazole regulates the blue copper active-site structure. J. Biol. Inorg. Chem. 12, 165–173 (2007)
Yanagisawa, S., Sato, K., Kikuchi, M., Kohzuma, T. & Dennison, C. Introduction of a π-π interaction at the active site of a cupredoxin: characterization of the Met16Phe pseudoazurin mutant. Biochemistry 42, 6853–6862 (2003)
Kanbi, L. D. et al. Crystal structures of the Met148 and Ser86Asp mutants of rusticyanin from Thiobacillus ferrooxidans: insights into the structural relationship with the cupredoxins and the multi copper proteins. J. Mol. Biol. 320, 263–275 (2002)
Hall, J. F., Kanbi, L. D., Harvey, I., Murphy, L. M. & Hasnain, S. S. Modulating the redox potential and acid stability of rusticyanin by site-directed mutagenesis of Ser86. Biochemistry 37, 11451–11458 (1998)
Grossmann, J. G., Ingledew, W. J., Harvey, I., Strange, R. W. & Hasnain, S. S. X-ray absorption studies and homology modeling define the structural features that specify the nature of the copper site in rusticyanin. Biochemistry 34, 8406–8414 (1995)
Walter, R. L. et al. Multiple wavelength anomalous diffraction (MAD) crystal structure of rusticyanin: a highly oxidizing cupredoxin with extreme acid stability. J. Mol. Biol. 263, 730–751 (1996)
Lowery, M. D. & Solomon, E. I. Axial ligand bonding in blue copper proteins. Inorg. Chim. Acta 198–200, 233–243 (1992)
Alcaraz, L. A. et al. Folding and unfolding in the blue copper protein rusticyanin: role of the oxidation state. Bioinorg. Chem. Appl. 1–9 (2007)
Hall, J. F., Kanbi, L. D., Strange, R. W. & Hasnain, S. S. Role of the axial ligand in type 1 Cu centers studied by point mutations of Met148 in rusticyanin. Biochemistry 38, 12675–12680 (1999)
Giudici-Orticoni, M. T., Guerlesquin, F., Bruschi, M. & Nitschke, W. Interaction-induced redox switch in the electron transfer complex rusticyanin-cytochrome c 4 . J. Biol. Chem. 274, 30365–30369 (1999)
Hart, P. J. et al. A missing link in cupredoxins: crystal structure of cucumber stellacyanin at 1.6 Å resolution. Protein Sci. 5, 2175–2183 (1996)
Romero, A. et al. X-ray analysis and spectroscopic characterization of M121Q azurin. A copper site model for stellacyanin. J. Mol. Biol. 229, 1007–1021 (1993)
Yanagisawa, S., Banfield, M. J. & Dennison, C. The role of hydrogen bonding at the active site of a cupredoxin: the Phe114Pro azurin variant. Biochemistry 45, 8812–8822 (2006)
Chang, T. K. et al. Gene synthesis, expression, and mutagenesis of the blue copper proteins azurin and plastocyanin. Proc. Natl Acad. Sci. USA 88, 1325–1329 (1991)
Mizoguchi, T. J., Di Bilio, A. J., Gray, H. B. & Richards, J. H. Blue to type 2 binding. Copper(II) and Cobalt(II) derivatives of a Cys112Asp mutant of Pseudomonas aeruginosa azurin. J. Am. Chem. Soc. 114, 10076–10078 (1992)
Jeuken, L. J. C. & Armstrong, F. A. Electrochemical origin of hysteresis in the electron-transfer reactions of adsorbed proteins: contrasting behavior of the “blue” copper protein, azurin, adsorbed on pyrolytic graphite and modified gold electrodes. J. Phys. Chem. B 105, 5271–5282 (2001)
Nilges, M. J. Electron Paramagnetic Resonance Studies of Low Symmetry Nickel(i) and Molybdenum(v) Complexes. Thesis, Univ. Illinois (1979)
Chang, H. R. et al. An unusually stable manganese(II)manganese(III) complex with novel EPR spectra: synthesis, structure, magnetism, and EPR analysis. J. Am. Chem. Soc. 110, 625–627 (1988)
Acknowledgements
This material is based on work supported by the US National Science Foundation under award no. CHE 05-52008. We thank E. Marshall, D. Poe, A. Huang and J. Li for discussions, and for help in protein expression and purification, and in spectroscopic and electrochemical data collection. N.M.M. is an NIH Predoctoral trainee, supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award 5 T32 GM070421 from the National Institute of General Medical Sciences.
Author Contributions N.M.M. performed most of the experimentation, led the study and authored most of the manuscript. D.K.G. and T.D.W. contributed intellectually and assisted in experimentation and editing. Crystal diffraction patterns were collected by H.R. and refined into PDB structures by Y.-G.G. M.J.N. assisted with EPR data collection and simulation. Y.L. designed and guided the project, and edited the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Crystallographic data have been deposited at the Protein Data Bank with the following PDB codes: N47S/M121L Az, 3JT2; N47S/F114N Az, 3JTB; F114P/M121Q Az, 3IN0.
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Supplementary Figures S1-12 with Legends, Supplementary Tables S1-S4 and Supplementary References. (PDF 453 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Marshall, N., Garner, D., Wilson, T. et al. Rationally tuning the reduction potential of a single cupredoxin beyond the natural range . Nature 462, 113–116 (2009). https://doi.org/10.1038/nature08551
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08551
This article is cited by
-
Assessing the role of redox partners in TthLPMO9G and its mutants: focus on H2O2 production and interaction with cellulose
Biotechnology for Biofuels and Bioproducts (2024)
-
The structure of plastocyanin tunes the midpoint potential by restricting axial ligation of the reduced copper ion
Communications Chemistry (2023)
-
Cross-talk between cancer and Pseudomonas aeruginosa mediates tumor suppression
Communications Biology (2023)
-
Orchestrating copper binding: structure and variations on the cupredoxin fold
JBIC Journal of Biological Inorganic Chemistry (2022)
-
A genetically encoded photosensitizer protein facilitates the rational design of a miniature photocatalytic CO2-reducing enzyme
Nature Chemistry (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.