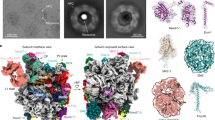
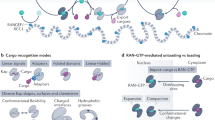
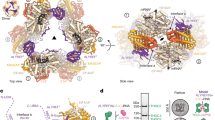
Abstract
Transfer RNAs are among the most ubiquitous molecules in cells, central to decoding information from messenger RNAs on translating ribosomes. In eukaryotic cells, tRNAs are actively transported from their site of synthesis in the nucleus to their site of function in the cytosol. This is mediated by a dedicated nucleo-cytoplasmic transport factor of the karyopherin-β family (Xpot, also known as Los1 in Saccharomyces cerevisiae). Here we report the 3.2 Å resolution structure of Schizosaccharomyces pombe Xpot in complex with tRNA and RanGTP, and the 3.1 Å structure of unbound Xpot, revealing both nuclear and cytosolic snapshots of this transport factor. Xpot undergoes a large conformational change on binding cargo, wrapping around the tRNA and, in particular, binding to the tRNA 5′ and 3′ ends. The binding mode explains how Xpot can recognize all mature tRNAs in the cell and yet distinguish them from those that have not been properly processed, thus coupling tRNA export to quality control.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kohler, A. & Hurt, E. Exporting RNA from the nucleus to the cytoplasm. Nature Rev. Mol. Cell Biol. 8, 761–773 (2007)
Rodriguez, M. S., Dargemont, C. & Stutz, F. Nuclear export of RNA. Biol. Cell 96, 639–655 (2004)
Elad, N., Maimon, T., Frenkiel-Krispin, D., Lim, R. Y. & Medalia, O. Structural analysis of the nuclear pore complex by integrated approaches. Curr. Opin. Struct. Biol. 19, 226–232 (2009)
Lim, R. Y., Aebi, U. & Fahrenkrog, B. Towards reconciling structure and function in the nuclear pore complex. Histochem. Cell Biol. 129, 105–116 (2008)
Peters, R. Translocation through the nuclear pore: Kaps pave the way. Bioessays 31, 466–477 (2009)
Zasloff, M. tRNA transport from the nucleus in a eukaryotic cell: carrier-mediated translocation process. Proc. Natl Acad. Sci. USA 80, 6436–6440 (1983)
Arts, G. J., Fornerod, M. & Mattaj, I. W. Identification of a nuclear export receptor for tRNA. Curr. Biol. 8, 305–314 (1998)
Kutay, U. et al. Identification of a tRNA-specific nuclear export receptor. Mol. Cell 1, 359–369 (1998)
Hellmuth, K. et al. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18, 6374–6386 (1998)
Cook, A., Bono, F., Jinek, M. & Conti, E. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671 (2007)
Madrid, A. S. & Weis, K. Nuclear transport is becoming crystal clear. Chromosoma 115, 98–109 (2006)
Calado, A., Treichel, N., Muller, E. C., Otto, A. & Kutay, U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 21, 6216–6224 (2002)
Shibata, S. et al. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 34, 4711–4721 (2006)
Jovine, L., Djordjevic, S. & Rhodes, D. The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: cleavage by Mg2+ in 15-year old crystals. J. Mol. Biol. 301, 401–414 (2000)
Shi, H. & Moore, P. B. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6, 1091–1105 (2000)
Marvin, M. C. & Engelke, D. R. RNase P: increased versatility through protein complexity? RNA Biol. 6, 40–42 (2009)
Spath, B., Canino, G. & Marchfelder, A. tRNase Z: the end is not in sight. Cell. Mol. Life Sci. 64, 2404–2412 (2007)
Xiong, Y. & Steitz, T. A. A story with a good ending: tRNA 3′-end maturation by CCA-adding enzymes. Curr. Opin. Struct. Biol. 16, 12–17 (2006)
Lund, E. & Dahlberg, J. E. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282, 2082–2085 (1998)
Bjork, G. R. et al. Transfer RNA modification. Annu. Rev. Biochem. 56, 263–287 (1987)
Iwata-Reuyl, D. An embarrassment of riches: the enzymology of RNA modification. Curr. Opin. Chem. Biol. 12, 126–133 (2008)
Arts, G. J., Kuersten, S., Romby, P., Ehresmann, B. & Mattaj, I. W. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 17, 7430–7441 (1998)
Lipowsky, G. et al. Coordination of tRNA nuclear export with processing of tRNA. RNA 5, 539–549 (1999)
Chook, Y. M. & Blobel, G. Structure of the nuclear transport complex karyopherin-β2–Ran·GppNHp. Nature 399, 230–237 (1999)
Lee, S. J., Matsuura, Y., Liu, S. M. & Stewart, M. Structural basis for nuclear import complex dissociation by RanGTP. Nature 435, 693–696 (2005)
Matsuura, Y. & Stewart, M. Structural basis for the assembly of a nuclear export complex. Nature 432, 872–877 (2004)
Monecke, T. et al. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science 324, 1087–1091 (2009)
Bischoff, F. R., Klebe, C., Kretschmer, J., Wittinghofer, A. & Ponstingl, H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl Acad. Sci. USA 91, 2587–2591 (1994)
Bloomfield, V. A., Crothers, D. N. & Tinoco, I. Nucleic Acids: Structures, Properties and Functions Ch. 8 298–300 (Macmillan, 1999)
Vetter, I. R., Arndt, A., Kutay, U., Gorlich, D. & Wittinghofer, A. Structural view of the Ran–Importin β interaction at 2.3 Å resolution. Cell 97, 635–646 (1999)
Andrade, M. A., Petosa, C., O’Donoghue, S. I., Muller, C. W. & Bork, P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 (2001)
Cingolani, G., Petosa, C., Weis, K. & Müller, C. W. Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229 (1999)
Kuersten, S., Arts, G. J., Walther, T. C., Englmeier, L. & Mattaj, I. W. Steady-state nuclear localization of exportin-t involves RanGTP binding and two distinct nuclear pore complex interaction domains. Mol. Cell. Biol. 22, 5708–5720 (2002)
Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A. & Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153 (1998)
Fukuhara, N., Fernandez, E., Ebert, J., Conti, E. & Svergun, D. Conformational variability of nucleo-cytoplasmic transport factors. J. Biol. Chem. 279, 2176–2181 (2004)
Cook, A. et al. The structure of the nuclear export receptor Cse1 in its cytosolic state reveals a closed conformation incompatible with cargo binding. Mol. Cell 18, 355–367 (2005)
Dong, X., Biswas, A. & Chook, Y. M. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nature Struct. Mol. Biol. 16, 558–560 (2009)
Moras, D. Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem. Sci. 17, 159–164 (1992)
LeMaster, D. M. & Richards, F. M. 1H–15N heteronuclear NMR studies of Escherichia coli thioredoxin in samples isotopically labeled by residue type. Biochemistry 24, 7263–7268 (1985)
Walker, P. A. et al. Efficient and rapid affinity purification of proteins using recombinant fusion proteases. Biotechnology 12, 601–605 (1994)
Doublie, S. et al. Crystallization and preliminary X-ray analysis of the 9 kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 384, 219–221 (1996)
Leslie, A. G. W. in Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography No. 26 (CCP4, 1992)
Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Broennimann et al. The PILATUS 1M detector. J. Synchrotron Radiat. 13, 120–130 (2006)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993)
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 (2002)
de La Fortelle, E. & Bricogne, G. in Macromolecular Crystallography Part A (eds Sweet, R. M. & Carter, C. W. J.) 472–494 (Academic, 1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Stein, N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Crystallogr. 41, 641–643 (2008)
Jones, T. A., Bergdoll, M. & Kjekdgaard, M. in Crystallographic and Modeling Methods in Molecular Design (eds Bugg, C. & Ealick, S.) 189–195 (Springer-Verlag, 1990)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004)
Landau, M. et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 (2005)
Acknowledgements
We would like to thank P. Brick for comments and critical reading of the manuscript. We also thank P. Reichelt and J. Ebert for technical assistance, S. Kuersten and I. Mattaj at the initial phase of the project, K. Valer-Saldana, S. Pleyer and J. Basquin of the MPI-Martinsried crystallization facility and the staff at the Swiss Light Source (Villigen, Switzerland) for assistance during data collection. We would like to thank C. Vonrhein (Global Phasing) for optimising the phase calculations of unbound Xpot with SHARP. This study was supported by the Max Planck Gesellschaft, the European Molecular Biology Laboratory (EMBL), the Sonderforschungsbereich SFB646 and the Gottfried Wilhelm Leibniz Program of the Deutsche Forschungsgemeinschaft (DFG), the EU grant 3D Repertoire contract number LSHG-CT-2005-512028.
Author Contributions N.F. identified S. pombe Xpot as the most promising orthologue for crystallisation, created the truncated tRNA construct and obtained initial, low-resolution crystals for the cargo-bound complex. M.J. crystallised unbound Xpot. A.G.C. solved the structure of unbound Xpot, obtained well-diffracting crystals of the ternary complex and solved its structure. E.C. supervised the project. A.G.C. and E.C. wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2 and Supplementary Figures 1-10 with Legends and Supplementary References for Figure 8. (PDF 10586 kb)
Rights and permissions
About this article
Cite this article
Cook, A., Fukuhara, N., Jinek, M. et al. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 461, 60–65 (2009). https://doi.org/10.1038/nature08394
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08394
This article is cited by
-
Karyopherin-mediated nucleocytoplasmic transport
Nature Reviews Molecular Cell Biology (2022)
-
Evolution and structural variations in chloroplast tRNAs in gymnosperms
BMC Genomics (2021)
-
Distinct mutations in importin-β family nucleocytoplasmic transport receptors transportin-SR and importin-13 affect specific cargo binding
Scientific Reports (2021)
-
Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A
Nature Communications (2016)
-
Precursors of tRNAs are stabilized by methylguanosine cap structures
Nature Chemical Biology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.