Abstract
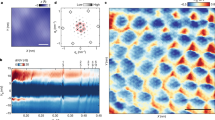
We live in a macroscopic three-dimensional (3D) world, but our best description of the structure of matter is at the atomic and molecular scale. Understanding the relationship between the two scales requires a bridge from the molecular world to the macroscopic world. Connecting these two domains with atomic precision is a central goal of the natural sciences, but it requires high spatial control of the 3D structure of matter1. The simplest practical route to producing precisely designed 3D macroscopic objects is to form a crystalline arrangement by self-assembly, because such a periodic array has only conceptually simple requirements: a motif that has a robust 3D structure, dominant affinity interactions between parts of the motif when it self-associates, and predictable structures for these affinity interactions. Fulfilling these three criteria to produce a 3D periodic system is not easy, but should readily be achieved with well-structured branched DNA motifs tailed by sticky ends2. Complementary sticky ends associate with each other preferentially and assume the well-known B-DNA structure when they do so3; the helically repeating nature of DNA facilitates the construction of a periodic array. It is essential that the directions of propagation associated with the sticky ends do not share the same plane, but extend to form a 3D arrangement of matter. Here we report the crystal structure at 4 Å resolution of a designed, self-assembled, 3D crystal based on the DNA tensegrity triangle4. The data demonstrate clearly that it is possible to design and self-assemble a well-ordered macromolecular 3D crystalline lattice with precise control.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Whitesides, G. M., Mathias, J. P. & Seto, C. T. Molecular self-assembly and nanochemistry: A chemical strategy for the synthesis of nanostructures. Science 254, 1312–1319 (1991)
Seeman, N. C. Nucleic acid junctions and crystal formation. J. Biomol. Struct. Dyn. 3, 11–34 (1985)
Qiu, H., Dewan, J. C. & Seeman, N. C. A DNA decamer with a sticky end: the crystal structure of d-CGACGATCGT. J. Mol. Biol. 267, 881–898 (1997)
Liu, D., Wang, W., Deng, Z., Walulu, R. & Mao, C. Tensegrity: construction of rigid DNA triangles with flexible four-arm junctions. J. Am. Chem. Soc. 126, 2324–2325 (2004)
Winfree, E., Liu, F., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998)
Nykypanchuk, D. M. M., Maye, M. M., van der Lelie, D. & Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 451, 549–552 (2008)
Park, S. Y. et al. DNA-programmable nanoparticle crystallization. Nature 451, 553–556 (2008)
Vargason, J. M., Henderson, K. & Ho, P. S. A crystallographic map of the transition from B-DNA to A-DNA. Proc. Natl Acad. Sci. USA 98, 7265–7270 (2001)
Eichmann, B. F., Vargason, J. M., Mooers, B. H. M. & Ho, P. S. The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions. Proc. Natl Acad. Sci. USA 97, 3971–3976 (2000)
Mao, C., Sun, W. & Seeman, N. C. Designed two-dimensional DNA Holliday junction arrays visualized by atomic force microscopy. J. Am. Chem. Soc. 121, 5437–5443 (1999)
Sha, R., Liu, F. & Seeman, N. C. Atomic force measurement of the inter-domain angle in symmetric Holliday junctions. Biochemistry 41, 5950–5955 (2002)
Birac, J. J., Sherman, W. B., Kopatsch, J., Constantinou, P. E. & Seeman, N. C. GIDEON, a program for design in structural DNA nanotechnology. J. Mol. Graph. Model. 25, 470–480 (2006)
Paukstelis, P. J., Nowakowski, J., Birktoft, J. J. & Seeman, N. C. The crystal structure of a continuous three-dimensional DNA lattice. Chem. Biol. 11, 1119–1126 (2004)
Paukstelis, P. J. Three dimensional DNA crystals as molecular sieves. J. Am. Chem. Soc. 128, 6794–6795 (2006)
Furukawa, H., Kim, H. J., Ockwig, N. W., O’Keeffe, M. & Yaghi, O. M. Control of vertex geometry, structure dimensionality, functionality, and pore metrics in the reticular synthesis of crystalline metal-organic frameworks and polyhedra. J. Am. Chem. Soc. 130, 11650–11651 (2008)
Kawano, M., Kawamichi, T., Haneda, T., Kajima, T. & Fujita, M. The modular synthesis of functional porous coordination networks. J. Am. Chem. Soc. 129, 15418–15419 (2007)
Ding, B. & Seeman, N. C. Operation of a DNA robot arm inserted into a 2D DNA crystalline substrate. Science 314, 1583–1585 (2006)
Zheng, J. et al. 2D nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 6, 1502–1504 (2006)
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982)
Robinson, B. H. & Seeman, N. C. The design of a biochip: a self-assembling molecular-scale memory device. Protein Eng. 1, 295–300 (1987)
Gu, H., Chao, J., Xiao, S.-J. & Seeman, N. C. Dynamic patterns programmed by DNA tiles captured on a DNA origami substrate. Nature Nanotech. 4, 245–249 (2009)
Rothemund, P. W. K., Papadakis, N. & Winfree, E. Algorithmic self-assembly of DNA Sierpinski triangles. PLoS Biol. 2, 2041–2053 (2004)
Seeman, N. C. De novo design of sequences for nucleic acid structure engineering. J. Biomol. Struct. Dyn. 8, 573–581 (1990)
Rosenbaum, G. et al. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J. Synchrotron Radiat. 13, 30–45 (2006)
Acknowledgements
This research has been supported by grants to N.C.S. from the National Institute of General Medical Sciences, the National Science Foundation, the Army Research Office, the Office of Naval Research and the W. M. Keck Foundation. It has also been supported by NSF grant CCF-0622093 and NIH grant 1R21EB007472 to C.M. We thank W. Sherman for assistance in establishing the likely structural features of tensegrity triangles. We thank R. Sweet, M. Allaire, H. Robinson, A. Saxena and A. Héroux at the BNL-NSLS at beamlines X6A and X25 of the National Synchrotron Light Source. BNL-NSLS is supported principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. The use of the 19ID beamline at the Structural Biology Center/Advanced Photon Source is supported by the US Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Author Contributions: J.Z. grew crystals, collected data, analysed data and wrote the paper; J.J.B. collected data, analysed data and wrote the paper; Y.C. grew crystals, collected data and analysed data; T.W. grew crystals, collected data, analysed data and wrote the paper; R.S. grew crystals, analysed data and wrote the paper; P.E.C. grew crystals and analysed data; S.L.G. collected data and analysed data; C.M. devised the motif, analysed data and wrote the paper; N.C.S. initiated the project, analysed data and wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Atomic coordinates and experimental structure factors have been deposited within the Protein Data Bank and are accessible under the code 3GBI.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Table S1 and Supplementary References. (PDF 199 kb)
Rights and permissions
About this article
Cite this article
Zheng, J., Birktoft, J., Chen, Y. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009). https://doi.org/10.1038/nature08274
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08274
This article is cited by
-
Exploring the diverse biomedical applications of programmable and multifunctional DNA nanomaterials
Journal of Nanobiotechnology (2023)
-
Multi-micron crisscross structures grown from DNA-origami slats
Nature Nanotechnology (2023)
-
Modular assembly of metal nanoparticles/mesoporous carbon two-dimensional nanosheets
NPG Asia Materials (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.