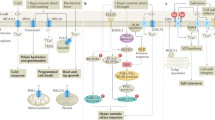
Abstract
Plant growth and development is regulated by a structurally unrelated collection of small molecules called plant hormones. During the last 15 years the number of known plant hormones has grown from five to at least ten. Furthermore, many of the proteins involved in plant hormone signalling pathways have been identified, including receptors for many of the major hormones. Strikingly, the ubiquitin–proteasome pathway plays a central part in most hormone-signalling pathways. In addition, recent studies confirm that hormone signalling is integrated at several levels during plant growth and development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Davies, P. J. in Plant Hormones: Physiology, Biochemistry and Molecular Biology (ed. Davies, P. J.) 1–12 (Kluwer Academic, 1995)
Grun, S., Lindermayr, C., Sell, S. & Durner, J. Nitric oxide and gene regulation in plants. J. Exp. Bot. 57, 507–516 (2006)
Vert, G., Nemhauser, J. L., Geldner, N., Hong, F. & Chory, J. Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21, 177–201 (2005)
Browse, J. Jasmonate: an oxylipin signal with many roles in plants. Vitam. Horm. 72, 431–456 (2005)
Loake, G. & Grant, M. Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472 (2007)
Gomez-Roldan, V. et al. Strigolactone inhibition of shoot branching. Nature 455, 189–194 (2008)
Umehara, M. et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 (2008)References 6 and 7 were the first papers to identify strigolactones as plant hormones that play a major part in inhibiting axillary bud outgrowth.
Jun, J. H., Fiume, E. & Fletcher, J. C. The CLE family of plant polypeptide signaling molecules. Cell. Mol. Life Sci. 65, 743–755 (2008)
Dharmasiri, N., Dharmasiri, S. & Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 (2005)
Dharmasiri, N. et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9, 109–119 (2005)
Kepinski, S. & Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 (2005)References 9 and 11 were the first papers to demonstrate that TIR1 was a receptor for auxin.
Ueguchi-Tanaka, M. et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698 (2005)
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007)
Thines, B. et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665 (2007)References 13 and 14 were the first to demonstrate that the JAZ proteins were substrates of COI1 and that jasmonate promoted their interactions. This breakthrough strongly suggested that COI1 was a receptor for jasmonate and that the signalling pathway was mechanistically similar to auxin perception and signalling.
Melotto, M. et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55, 979–988 (2008)
Pandey, S., Nelson, D. C. & Assmann, S. M. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis . Cell 136, 136–148 (2009)This paper identified two novel G-proteins that directly bind abscisic acid.
Chow, B. & McCourt, P. Plant hormone receptors: perception is everything. Genes Dev. 20, 1998–2008 (2006)
Schwechheimer, C. & Willige, B. C. Shedding light on gibberellic acid signalling. Curr. Opin. Plant Biol. 12, 57–62 (2009)
Rensing, S. A. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008)
Vandenbussche, F., Fierro, A. C., Wiedemann, G., Reski, R. & Van Der Straeten, D. Evolutionary conservation of plant gibberellin signalling pathway components. BMC Plant Biol. 7, 65 (2007)
Zhao, Y. The role of local biosynthesis of auxin and cytokinin in plant development. Curr. Opin. Plant Biol. 11, 16–22 (2008)
Mockaitis, K. & Estelle, M. Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24, 55–80 (2008)
Wasternack, C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100, 681–697 (2007)
Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251 (2008)
Hirano, K., Ueguchi-Tanaka, M. & Matsuoka, M. GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 13, 192–199 (2008)
Hirose, N. et al. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59, 75–83 (2008)
Symons, G. M., Ross, J. J., Jager, C. E. & Reid, J. B. Brassinosteroid transport. J. Exp. Bot. 59, 17–24 (2008)
Hirayama, T. & Shinozaki, K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12, 343–351 (2007)
Dharmasiri, S. & Estelle, M. The role of regulated protein degradation in auxin response. Plant Mol. Biol. 49, 401–409 (2002)
Moon, J., Parry, G. & Estelle, M. The ubiquitin-proteasome pathway and plant development. Plant Cell 16, 3181–3195 (2004)
Ruegger, M. et al. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207 (1998)
Gray, W. M., Kepinski, S., Rouse, D., Leyser, O. & Estelle, M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276 (2001)
Guilfoyle, T. J. & Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460 (2007)
Reed, J. W. Roles and activities of Aux/IAA proteins in Arabidopsis . Trends Plant Sci. 6, 420–425 (2001)
Dharmasiri, N., Dharmasiri, S., Jones, A. M. & Estelle, M. Auxin action in a cell-free system. Curr. Biol. 13, 1418–1422 (2003)
Kepinski, S. & Leyser, O. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc. Natl Acad. Sci. USA 101, 12381–12386 (2004)
Tan, X. et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 (2007)This paper described the crystal structure of the TIR1 auxin receptor in complex with auxin and its substrate. The structure has substantially improved our understanding of auxin perception.
Tan, X. & Zheng, N. Hormone signaling through protein destruction: a lesson from plants. Am. J. Physiol. Endocrinol. Metab. 296, E223–E227 (2009)
Hayashi, K. et al. Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl Acad. Sci. USA 105, 5632–5637 (2008)
Tiwari, S. B., Hagen, G. & Guilfoyle, T. J. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543 (2004)
Szemenyei, H., Hannon, M. & Long, J. A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 (2008)This paper demonstrated that TOPLESS directly binds domain I of Aux/IAA proteins, leading to transcriptional repression of auxin-responsive genes. This mechanism probably accounts for the repression of ARF function by Aux/IAA proteins.
Feys, B., Benedetti, C. E., Penfold, C. N. & Turner, J. G. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759 (1994)
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M. & Turner, J. G. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094 (1998)
Yan, Y. et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483 (2007)
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y. & Howe, G. A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl Acad. Sci. USA 105, 7100–7105 (2008)
de Lucas, M. et al. A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484 (2008)
Feng, S. et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479 (2008)References 46 and 47 revealed that the basic helix–loop–helix transcription factors PIF3 and PIF4 directly interact with DELLA proteins. Upon gibberellin accumulation, the DELLA proteins are destabilized, freeing PIF3 and PIF4 to regulate their target genes. This is a new and compelling model for DELLA-mediated growth regulation.
McGinnis, K. M. et al. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15, 1120–1130 (2003)
Sasaki, A. et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898 (2003)
Nakajima, M. et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46, 880–889 (2006)
Griffiths, J. et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis . Plant Cell 18, 3399–3414 (2006)
Willige, B. C. et al. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis . Plant Cell 19, 1209–1220 (2007)
Murase, K., Hirano, Y., Sun, T. P. & Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 (2008)
Shimada, A. et al. Structural basis for gibberellin recognition by its receptor GID1. Nature 456, 520–523 (2008)References 53 and 54 are the first to describe the structure of the GID1 gibberellin receptor.
Shen, Y. Y. et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443, 823–826 (2006)
Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A. & Chory, J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl Acad. Sci. USA 98, 2053–2058 (2001)
Muller, A. H. & Hansson, M. The barley magnesium chelatase 150-kDa subunit is not an abscisic-acid receptor. Plant Physiol. 150, 157–166 (2009)
Liu, X. et al. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315, 1712–1716 (2007)
Johnston, C. A. et al. Comment on “A G protein coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid”. Science 318, 914; author reply 914 (2007)
Guo, J., Zeng, Q., Emami, M., Ellis, B. E. & Chen, J. G. The GCR2 gene family is not required for ABA control of seed germination and early seedling development in Arabidopsis . PLoS One 3, e2982 (2008)
Gao, Y. et al. Genetic characterization reveals no role for the reported ABA receptor, GCR2, in ABA control of seed germination and early seedling development in Arabidopsis . Plant J. 52, 1001–1013 (2007)
Risk, J. M., Day, C. L. & Macknight, R. C. Re-evaluation of abscisic acid (ABA) binding assays shows that GCR2 does not bind ABA. Plant Physiol. 150, 6–11 (2009)
Park, S. Y. et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science Epub ahead of print, 10.1126/science.1173041 (30 April 2009)
Ma, Y. et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science Epub ahead of print, 10.1126/science.1172408 (8 May 2009)References 63 and 64 describe the identification of soluble abscisic-acid receptors. A novel model for abscisic-acid action is proposed, in which abscisic acid acts to inhibit PP2C proteins such as ABI1 and ABI2, by promoting an interaction between the phosphatase and PYR1/RCAR1 and related proteins.
McCourt, P. & Creelman, R. The ABA receptors—we report, you decide. Curr. Opin. Plant Biol. 11, 474–478 (2008)
Zhang, X., Garreton, V. & Chua, N. H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532–1543 (2005)
Stone, S. L., Williams, L. A., Farmer, L. M., Vierstra, R. D. & Callis, J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428 (2006)
Solano, R., Stepanova, A., Chao, Q. & Ecker, J. R. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714 (1998)
Guo, H. & Ecker, J. R. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677 (2003)
Potuschak, T. et al. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689 (2003)References 67 and 68 showed that the F-box proteins EBF1 and EBF2 were important regulators of EIN3 accumulation. The data demonstrate the pivital role of the ubiquitin-mediated degradation during ethylene signalling.
Binder, B. M. et al. The Arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19, 509–523 (2007)
Qiao, H., Chang, K. N., Yazaki, J. & Ecker, J. R. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis . Genes Dev. 23, 512–521 (2009)
Ongaro, V. & Leyser, O. Hormonal control of shoot branching. J. Exp. Bot. 59, 67–74 (2008)
Stirnberg, P., van De Sande, K. & Leyser, H. M. MAX1 and MAX2 control shoot lateral branching in Arabidopsis . Development 129, 1131–1141 (2002)
Johnson, X. et al. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol. 142, 1014–1026 (2006)
Wilson, A. K., Pickett, F. B., Turner, J. C. & Estelle, M. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222, 377–383 (1990)
Roman, G., Lubarsky, B., Kieber, J. J., Rothenberg, M. & Ecker, J. R. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics 139, 1393–1409 (1995)
Stepanova, A. N., Hoyt, J. M., Hamilton, A. A. & Alonso, J. M. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis . Plant Cell 17, 2230–2242 (2005)
Stepanova, A. N. et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 (2008)
Tao, Y. et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176 (2008)
Tsuchisaka, A. & Theologis, A. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 136, 2982–3000 (2004)
Tsuchisaka, A. & Theologis, A. Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc. Natl Acad. Sci. USA 101, 2275–2280 (2004)
Nagpal, P. et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107–4118 (2005)
Feraru, E. & Friml, J. PIN polar targeting. Plant Physiol. 147, 1553–1559 (2008)
Laplaze, L. et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19, 3889–3900 (2007)
Blilou, I. et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44 (2005)
Nemhauser, J. L., Mockler, T. C. & Chory, J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis . PLoS Biol. 2, E258 (2004)
Goda, H. et al. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis . Plant Physiol. 134, 1555–1573 (2004)
Mouchel, C. F., Osmont, K. S. & Hardtke, C. S. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443, 458–461 (2006)
Vert, G., Walcher, C. L., Chory, J. & Nemhauser, J. L. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl Acad. Sci. USA 105, 9829–9834 (2008)This paper elucidated an interesting molecular crosstalk strategy in which the brassinosteroid-regulated BIN2 kinase directly regulates ARF2, thereby modulating auxin signalling.
Weiss, D. & Ori, N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144, 1240–1246 (2007)
Fu, X. & Harberd, N. P. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421, 740–743 (2003)
Achard, P., Vriezen, W. H., Van Der Straeten, D. & Harberd, N. P. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15, 2816–2825 (2003)
Achard, P. et al. Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94 (2006)
Navarro, L. et al. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18, 650–655 (2008)
Acknowledgements
Work in M.E.’s laboratory was supported by grants from the NIH (GM43644), the NSF (IOS 0744800), and the DOE (DOE DE-FG02-02ER15312) to M.E.
Author Contributions A.S. and M.E. generated an outline together. A.S. prepared the first draft of the article and A.S. and M.E. worked together on all subsequent drafts.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Santner, A., Estelle, M. Recent advances and emerging trends in plant hormone signalling. Nature 459, 1071–1078 (2009). https://doi.org/10.1038/nature08122
Issue Date:
DOI: https://doi.org/10.1038/nature08122
This article is cited by
-
An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity
Nature Chemical Biology (2024)
-
Ethylene may be the Key Factor Leading to the Homologous Transformation of Stamens into Pistils in Three-Pistil Wheat
Journal of Plant Growth Regulation (2024)
-
Study on physiological and biochemical characteristics during in vitro flowering of Rosa ‘Yametsu-Hime’
Plant Cell, Tissue and Organ Culture (PCTOC) (2024)
-
Gut-Brain Axis Modulation of Metabolic Disorders: Exploring the Intertwined Neurohumoral Pathways and Therapeutic Prospects
Neurochemical Research (2024)
-
Differentially expression analyses in fruit of cultivated and wild species of grape and peach
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.