Abstract
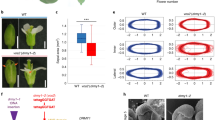
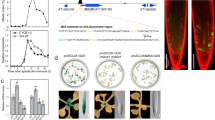
Local hormone maxima are essential for the development of multicellular structures and organs. For example, steroid hormones accumulate in specific cell types of the animal fetus to induce sexual differentiation1 and concentration peaks of the plant hormone auxin direct organ initiation and mediate tissue patterning2,3,4. Here we provide an example of a regulated local hormone minimum required during organogenesis. Our results demonstrate that formation of a local auxin minimum is necessary for specification of the valve margin separation layer where Arabidopsis fruit opening takes place. Consequently, ectopic production of auxin, specifically in valve margin cells, leads to a complete loss of proper cell fate determination. The valve margin identity factor INDEHISCENT (IND) is responsible for forming the auxin minimum by coordinating auxin efflux in separation-layer cells. We propose that the simplicity of formation and maintenance make local hormone minima particularly well suited to specify a small number of cells such as the stripes at the valve margins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wu, Z., Wan, S. & Lee, M. M. Key factors in the regulation of fetal and postnatal leydig cell development. J. Cell. Physiol. 213, 429–433 (2007)
Sabatini, S. et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472 (1999)
Friml, J. et al. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis . Nature 426, 147–153 (2003)
Benková, E. et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003)
Konishi, S. et al. An SNP caused loss of shattering during rice domestication. Science 312, 1392–1396 (2006)
Østergaard, L., Kempin, S. A., Bies, D., Klee, H. J. & Yanofsky, M. F. Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. Plant Biotechnol. J. 4, 45–51 (2006)
Roeder, A. H. K. & Yanofsky, M. F. in The Arabidopsis Book (eds Somerville, C. R. & Meyerowitz, E. M.) doi:10.1199/tab.0075 (Am. Soc. Plant Biol., 2006)
Petersen, M. et al. Isolation and characterisation of a pod dehiscence zone-specific polygalacturonase from Brassica napus . Plant Mol. Biol. 31, 517–527 (1996)
Nemhauser, J., Feldmann, L. J. & Zambryski, P. C. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888 (2000)
Østergaard, L. Don’t ‘leaf’ now. The making of a fruit. Curr. Opin. Plant Biol. 12, 36–41 (2009)
Chauvaux, N. et al. The role of auxin in cell separation in the dehiscence zone of rapeseed pods. J. Exp. Biol. 48, 1423–1429 (1997)
Sohlberg, J. J. et al. STY1 regulates auxin homeostasis and affects apical–basal patterning of the Arabidopsis gynoecium. Plant J. 47, 112–123 (2006)
Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. Early flower development in Arabidopsis . Plant Cell 2, 755–767 (1990)
Liljegren, S. J. et al. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116, 843–853 (2004)
Ferrándiz, C., Liljegren, S. J. & Yanofsky, M. F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289, 436–438 (2000)
Roeder, A. H., Ferrándiz, C. & Yanofsky, M. F. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13, 1630–1635 (2003)
Romano, C. P., Robson, P. R., Smith, H., Estelle, M. & Klee, H. Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6–1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 27, 1071–1083 (1995)
Aoyama, T. & Chua, N.-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612 (1997)
Gremski, K., Ditta, G. & Yanofsky, M. F. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana . Development 134, 3593–3601 (2007)
Okada, K., Ueda, J., Komaki, M. K., Bell, C. J. & Shimura, Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684 (1991)
Bennett, S. M. R., Alvarez, J., Bossinger, G. & Smyth, D. R. Morphogenesis in pinoid mutants of Arabidopsis thaliana . Plant J. 8, 505–520 (1995)
Petrásek, J. et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914–918 (2006)
Wisniewska, J. et al. PIN localization directs auxin flow in plants. Science 312, 883 (2006)
Tanaka, H., Dhonukshe, P., Brewer, P. B. & Friml, J. Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cell. Mol. Life Sci. 63, 2738–2754 (2006)
Benjamins, R., Quint, A., Weijers, D., Hooykaas, P. & Offringa, R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128, 4057–4067 (2001)
Friml, J. et al. A PINOID-dependent binary switch in apical–basal PIN polar targeting directs auin efflux. Science 306, 862–865 (2004)
Friml, J., Wisniewska, J., Benková, E., Mendgen, K. & Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis . Nature 415, 806–809 (2002)
Michniewicz, M. et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flow. Cell 130, 1044–1056 (2007)
Santner, A. A. & Watson, J. C. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis . Plant J. 45, 752–764 (2006)
Galván-Ampudia, C. S. & Offringa, R. Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 12, 541–547 (2007)
Yu, H., Ito, T., Wellmer, F. & Meyerowitz, E. M. Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nature Genet. 36, 157–161 (2004)
Cutler, S. R., Ehrhardt, D. W., Griffitts, J. S. & Somerville, C. R. Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl Acad. Sci. USA 97, 3718–3723 (2000)
Romano, C. P., Hein, M. B. & Klee, H. J. Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi . Genes Dev. 5, 438–446 (1991)
Edlund, A., Eklöf, S., Sundberg, B., Moritz, T. & Sandberg, G. A microscale technoque for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol. 108, 1043–1047 (1995)
Rozen, S. & Skaletsky, H. J. in Bioinformatics Methods and Protocols: Methods in Molecular Biology (eds Krawetz, S. & Misener, S.) 365–386 (Humana, 2000)
Han, S. & Kim, D. AtRTPrimer: database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC Bioinformatics 7, 179 (2006)
Morohashi, K. et al. Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145, 736–746 (2007)
Ito, T., Ng, K. H., Lim, T. S., Yu, H. & Meyerowitz, E. M. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis . Plant Cell 19, 3516–3529 (2007)
Blázquez, M. A., Soowal, L. N., Lee, I. & Weigel, D. LEAFY expression and flower initiation in Arabidopsis . Development 124, 3835–3844 (1997)
Acknowledgements
We thank E. York and K. Findlay for assistance on SEM analysis, G. Calder for assistance with confocal microscopy, P. Pople for graphics assistance, and H. F. Klee for the pMON518 plasmid containing the iaaM gene. We also wish to thank G. S. Ditta, L. Dolan, S. Fuentes, J. Kleine-Vehn, R. Sablowski, P. Stephenson and T. Wood for carefully reading the manuscript and constructive criticism. P.R was the recipient of a postdoctoral fellowship from the Spanish government. This work was supported by grants from FWO (Odysseus program) to J.F., from The Netherlands Organisation for Scientific Research (ALW-NWO) to R.O., from the National Science Foundation to M.F.Y., and from the Biotechnological and Biological Sciences Research Council as well as core strategic funds from the John Innes Centre to L.Ø.
Author Contributions This project was conceived by K.S., M.F.Y. and L.Ø. Experiments were designed by K.S. and L.Ø. K.S. performed the confocal microscopy and expression analyses, T.G. carried out the chromatin immunoprecipitations, L.Ø. made the IND::IND:iaaM lines and performed plant sections and tissue staining, K.L. performed the IAA measurements, P.R. made the 35S::IND:GR transgenic line, S.J.L. characterized the 35S::IND lines and took the 35S::IND SEM image shown, and C.S.G.-A., J.F. and R.O. analysed the effect of WAG2 activity on PIN localization. K.S. and L.Ø. analysed the data and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Data, a Supplementary Reference, Supplementary Figures 1-7 with Legends and Supplementary Notes. (PDF 7297 kb)
Rights and permissions
About this article
Cite this article
Sorefan, K., Girin, T., Liljegren, S. et al. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459, 583–586 (2009). https://doi.org/10.1038/nature07875
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07875
This article is cited by
-
Effect of ion source polarity and dopants on the detection of auxin plant hormones by ion mobility-mass spectrometry
Analytical and Bioanalytical Chemistry (2022)
-
Current overview on the genetic basis of key genes involved in soybean domestication
aBIOTECH (2022)
-
Coordination of biradial-to-radial symmetry and tissue polarity by HD-ZIP II proteins
Nature Communications (2021)
-
Seed-specific expression of TaYUC10 significantly increases auxin and protein content in wheat seeds
Plant Cell Reports (2021)
-
Down-regulation of MANNANASE7 gene in Brassica napus L. enhances silique dehiscence-resistance
Plant Cell Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.