Abstract
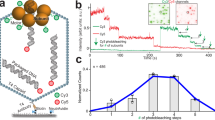
Homomeric ring ATPases perform many vital and varied tasks in the cell, ranging from chromosome segregation to protein degradation. Here we report the direct observation of the intersubunit coordination and step size of such a ring ATPase, the double-stranded-DNA packaging motor in the bacteriophage ϕ29. Using high-resolution optical tweezers, we find that packaging occurs in increments of 10 base pairs (bp). Statistical analysis of the preceding dwell times reveals that multiple ATPs bind during each dwell, and application of high force reveals that these 10-bp increments are composed of four 2.5-bp steps. These results indicate that the hydrolysis cycles of the individual subunits are highly coordinated by means of a mechanism novel for ring ATPases. Furthermore, a step size that is a non-integer number of base pairs demands new models for motor–DNA interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Latterich, M. & Patel, S. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 8, 65–71 (1998)
Ogura, T. & Wilkinson, A. J. AAA+ superfamily ATPases: common structure–diverse function. Genes Cells 6, 575–597 (2001)
Iyer, L. M., Leipe, D. D., Koonin, E. V. & Aravind, L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31 (2004)
Kainov, D. E., Tuma, R. & Mancini, E. J. Hexameric molecular motors: P4 packaging ATPase unravels the mechanism. Cell. Mol. Life Sci. 63, 1095–1105 (2006)
Erzberger, J. P. & Berger, J. M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 (2006)
Singleton, M. R., Sawaya, M. R., Ellenberger, T. & Wigley, D. B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101, 589–600 (2000)
Mancini, E. J. et al. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell 118, 743–755 (2004)
Kinosita, K., Adachi, K. & Itoh, H. Rotation of F1-ATPase: How an ATP-driven molecular machine may work. Annu. Rev. Biophys. Biomol. Struct. 33, 245–268 (2004)
Enemark, E. J. & Joshua-Tor, L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275 (2006)
Skordalakes, E. & Berger, J. M. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell 127, 553–564 (2006)
Adelman, J. L. et al. Mechanochemistry of transcription termination factor Rho. Mol. Cell 22, 611–621 (2006)
Liao, J.-C., Jeong, Y.-J., Kim, D.-E., Patel, S. S. & Oster, G. Mechanochemistry of T7 DNA helicase. J. Mol. Biol. 350, 452–475 (2005)
Massey, T. H., Mercogliano, C. P., Yates, J., Sherratt, D. J. & Löwe, J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell 23, 457–469 (2006)
Crampton, D. J., Mukherjee, S. & Richardson, C. C. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol. Cell 21, 165–174 (2006)
Gai, D., Zhao, R., Li, D., Finkielstein, C. V. & Chen, X. S. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119, 47–60 (2004)
Martin, A., Baker, T. A. & Sauer, R. T. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature 437, 1115–1120 (2005)
Guo, P., Grimes, S. & Anderson, D. A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage ϕ29. Proc. Natl Acad. Sci. USA 83, 3505–3509 (1986)
Smith, D. E. et al. The bacteriophage straight ϕ29 portal motor can package DNA against a large internal force. Nature 413, 748–752 (2001)
Chemla, Y. R. et al. Mechanism of force generation of a viral DNA packaging motor. Cell 122, 683–692 (2005)
Grimes, S., Jardine, P. J. & Anderson, D. Bacteriophage ϕ29 DNA packaging. Adv. Virus Res. 58, 255–294 (2002)
Hugel, T. et al. Experimental test of connector rotation during DNA packaging into bacteriophage ϕ29 capsids. PLoS Biol. 5, e59 (2007)
Fuller, D. N. et al. Ionic effects on viral DNA packaging and portal motor function in bacteriophage ϕ29. Proc. Natl Acad. Sci. USA 104, 11245–11250 (2007)
Rickgauer, J. P. et al. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage ϕ29. Biophys. J. 94, 159–167 (2008)
Simpson, A. A. et al. Structure of the bacteriophage ϕ29 DNA packaging motor. Nature 408, 745–750 (2000)
Morais, M. C. et al. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage ϕ29. J. Struct. Biol. 135, 38–46 (2001)
Morais, M. C. et al. Defining molecular and domain boundaries in the bacteriophage ϕ29 DNA packaging motor. Structure 16, 1267–1274 (2008)
Burroughs, A. M., Iyer, L. M. & Aravind, L. in Gene and Protein Evolution (ed. Volff, J.-N.) 48–65 (Karger, 2007)
Iyer, L. M., Makarova, K. S., Koonin, E. V. & Aravind, L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 32, 5260–5279 (2004)
Guo, P., Peterson, C. & Anderson, D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage ϕ29. J. Mol. Biol. 197, 229–236 (1987)
Chen, C. & Guo, P. Sequential action of six virus-encoded DNA-packaging RNAs during phage ϕ29 genomic DNA translocation. J. Virol. 71, 3864–3871 (1997)
Moffitt, J. R., Chemla, Y. R., Smith, S. B. & Bustamante, C. Recent advances in optical tweezers. Annu. Rev. Biochem. 77, 205–228 (2008)
Moffitt, J. R., Chemla, Y. R., Izhaky, D. & Bustamante, C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl Acad. Sci. USA 103, 9006–9011 (2006)
Bustamante, C., Chemla, Y. R. & Moffitt, J. R. in Single-Molecule Techniques: A Laboratory Manual (eds Selvin, P. R. & Ha, T.) 297–324 (Cold Spring Harbor Laboratories, 2008)
Schnitzer, M. J. & Block, S. M. Statistical kinetics of processive enzymes. Cold Spring Harb. Symp. Quant. Biol. 60, 793–802 (1995)
Koza, Z. Maximal force exerted by a molecular motor. Phys. Rev. E 65, 031905 (2002)
Chemla, Y. R., Moffitt, J. R. & Bustamante, C. Exact solutions for kinetic models of macromolecular dynamics. J. Phys. Chem. B 112, 6025–6044 (2008)
Bustamante, C., Chemla, Y. R., Forde, N. R. & Izhaky, D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 73, 705–748 (2004)
Segel, I. H. Enzyme Kinetics (John Wiley & Sons, 1975)
Oster, G. & Wang, H. Reverse engineering a protein: the mechanochemistry of ATP synthase. Biochim. Biophys. Acta 1458, 482–510 (2000)
Skordalakes, E. & Berger, J. M. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell 114, 135–146 (2003)
Lisal, J. et al. Functional visualization of viral molecular motor by hydrogen-deuterium exchange reveals transient states. Nature Struct. Mol. Biol. 12, 460–466 (2005)
Moreau, M. J., McGeoch, A. T., Lowe, A. R., Itzhaki, L. S. & Bell, S. D. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol. Cell 28, 304–314 (2007)
Berg-Sorensen, K. & Flyvbjerg, H. Power spectrum analysis for optical tweezers. Rev. Sci. Instrum. 75, 594–612 (2004)
Block, S. M. & Svoboda, K. Analysis of high resolution recordings of motor movement. Biophys. J. 68, 230–241 (1995)
Carter, N. J. & Cross, R. A. Mechanics of the kinesin step. Nature 435, 308–312 (2005)
Parzen, E. On estimation of a probability density function and mode. Ann. Math. Stat. 33, 1065–1076 (1962)
Grimes, S. & Anderson, D. The bacteriophage ϕ29 packaging proteins supercoil the DNA ends. J. Mol. Biol. 266, 901–914 (1997)
Kellner, L. The near infra-red absorption spectrum of heavy water. Proc. R. Soc. Lond. A 159, 0410–0415 (1937)
Baumann, C. G., Smith, S. B., Bloomfield, V. A. & Bustamante, C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl Acad. Sci. USA 94, 6185–6190 (1997)
Yanagi, K., Prive, G. G. & Dickerson, R. E. Analysis of local helix geometry in three B-DNA decamers and eight dodecamers. J. Mol. Biol. 217, 201–214 (1991)
Acknowledgements
We thank C. L. Hetherington, M. Nollmann and G. Chistol for a critical reading of the manuscript; C. L. Hetherington, A. Politzer, M. Strycharska, M. Kopaczynska and J. Yu for critical discussions; and J. Choy, S. Grill and S. Smith for advice regarding instrumentation. J.R.M. acknowledges the National Science Foundation’s Graduate Research Fellowship and Y.R.C. the Burroughs Welcome Fund’s Career Awards at the Scientific Interface for funding. This research was supported in part by NIH grants GM-071552, DE-003606 and GM-059604. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Contributions J.R.M., Y.R.C. and K.A. conducted the experiments and performed the analysis; S.G., P.J.J. and D.L.A. prepared and provided experimental materials; and J.R.M., Y.R.C., K.A., S.G., P.J.J., D.L.A. and C.B. wrote the paper. J.R.M. and Y.R.C. contributed equally to this work.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Methods, a Supplementary Discussion, Supplementary Figures 1-8 with Legends and Supplementary References (PDF 1112 kb)
Rights and permissions
About this article
Cite this article
Moffitt, J., Chemla, Y., Aathavan, K. et al. Intersubunit coordination in a homomeric ring ATPase. Nature 457, 446–450 (2009). https://doi.org/10.1038/nature07637
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07637
This article is cited by
-
Modelling the Effect of Geometry and Loading on Mechanical Response of SARS-CoV-2
BioNanoScience (2022)
-
Single-molecule force spectroscopy reveals the dynamic strength of the hair-cell tip-link connection
Nature Communications (2021)
-
Extreme parsimony in ATP consumption by 20S complexes in the global disassembly of single SNARE complexes
Nature Communications (2021)
-
Optical tweezers in single-molecule biophysics
Nature Reviews Methods Primers (2021)
-
A DNA packaging motor inchworms along one strand allowing it to adapt to alternative double-helical structures
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.