Abstract
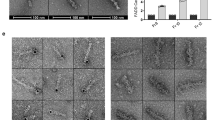
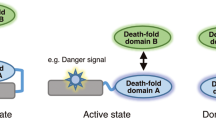
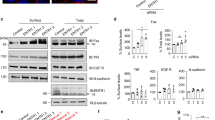
The death inducing signalling complex (DISC) formed by Fas receptor, FADD (Fas-associated death domain protein) and caspase 8 is a pivotal trigger of apoptosis1,2,3. The Fas–FADD DISC represents a receptor platform, which once assembled initiates the induction of programmed cell death. A highly oligomeric network of homotypic protein interactions comprised of the death domains of Fas and FADD is at the centre of DISC formation4,5. Thus, characterizing the mechanistic basis for the Fas–FADD interaction is crucial for understanding DISC signalling but has remained unclear largely because of a lack of structural data. We have successfully formed and isolated the human Fas–FADD death domain complex and report the 2.7 Å crystal structure. The complex shows a tetrameric arrangement of four FADD death domains bound to four Fas death domains. We show that an opening of the Fas death domain exposes the FADD binding site and simultaneously generates a Fas–Fas bridge. The result is a regulatory Fas–FADD complex bridge governed by weak protein–protein interactions revealing a model where the complex itself functions as a mechanistic switch. This switch prevents accidental DISC assembly, yet allows for highly processive DISC formation and clustering upon a sufficient stimulus. In addition to depicting a previously unknown mode of death domain interactions, these results further uncover a mechanism for receptor signalling solely by oligomerization and clustering events.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Ashkenazi, A. & Dixit, V. M. Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 11, 255–260 (1999)
Peter, M. E. & Krammer, P. H. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 10, 26–35 (2003)
Taylor, R. C., Cullen, S. P. & Martin, S. J. Apoptosis: controlled demolition at the cellular level. Nature Rev. Mol. Cell Biol. 9, 231–241 (2008)
Fesik, S. W. Insights into programmed cell death through structural biology. Cell 103, 273–282 (2000)
Park, H. H. et al. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu. Rev. Immunol. 25, 561–586 (2007)
Festjens, N., Cornelis, S., Lamkanfi, M. & Vandenabeele, P. Caspase-containing complexes in the regulation of cell death and inflammation. Biol. Chem. 387, 1005–1016 (2006)
Tibbetts, M. D., Zheng, L. & Lenardo, M. J. The death effector domain protein family: regulators of cellular homeostasis. Nature Immunol. 4, 404–409 (2003)
Algeciras-Schimnich, A. et al. Molecular ordering of the initial signaling events of CD95. Mol. Cell. Biol. 22, 207–220 (2002)
Feig, C., Tchikov, V., Schutze, S. & Peter, M. E. Palmitoylation of CD95 facilitates formation of SDS-stable receptor aggregates that initiate apoptosis signaling. EMBO J. 26, 221–231 (2007)
Lavrik, I. N. et al. Analysis of CD95 threshold signaling: triggering of CD95 (FAS/APO-1) at low concentrations primarily results in survival signaling. J. Biol. Chem. 282, 13664–13671 (2007)
Muppidi, J. R. & Siegel, R. M. Ligand-independent redistribution of Fas (CD95) into lipid rafts mediates clonotypic T cell death. Nature Immunol. 5, 182–189 (2004)
O’Reilly, L. A. et al. Modifications and intracellular trafficking of FADD/MORT1 and caspase-8 after stimulation of T lymphocytes. Cell Death Differ. 11, 724–736 (2004)
Siegel, R. M. et al. SPOTS: signaling protein oligomeric transduction structures are early mediators of death receptor-induced apoptosis at the plasma membrane. J. Cell Biol. 167, 735–744 (2004)
Werner, M. H., Wu, C. & Walsh, C. M. Emerging roles for the death adaptor FADD in death receptor avidity and cell cycle regulation. Cell Cycle 5, 2332–2338 (2006)
Petrilli, V., Dostert, C., Muruve, D. A. & Tschopp, J. The inflammasome: a danger sensing complex triggering innate immunity. Curr. Opin. Immunol. 19, 615–622 (2007)
Riedl, S. J. & Salvesen, G. S. The apoptosome: signalling platform of cell death. Nature Rev. Mol. Cell Biol. 8, 405–413 (2007)
Berglund, H. et al. The three-dimensional solution structure and dynamic properties of the human FADD death domain. J. Mol. Biol. 302, 171–188 (2000)
Carrington, P. E. et al. The structure of FADD and its mode of interaction with procaspase-8. Mol. Cell 22, 599–610 (2006)
Jeong, E. J. et al. The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD. J. Biol. Chem. 274, 16337–16342 (1999)
Huang, B., Eberstadt, M., Olejniczak, E. T., Meadows, R. P. & Fesik, S. W. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature 384, 638–641 (1996)
Clackson, T. & Wells, J. A. A hot spot of binding energy in a hormone-receptor interface. Science 267, 383–386 (1995)
Reichmann, D., Rahat, O., Cohen, M., Neuvirth, H. & Schreiber, G. The molecular architecture of protein-protein binding sites. Curr. Opin. Struct. Biol. 17, 67–76 (2007)
Bogan, A. A. & Thorn, K. S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998)
Huxford, T. et al. Solvent exposed non-contacting amino acids play a critical role in NF-κB/IκBα complex formation. J. Mol. Biol. 324, 587–597 (2002)
Janin, J., Miller, S. & Chothia, C. Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol. 204, 155–164 (1988)
Roisman, L. C., Jaitin, D. A., Baker, D. P. & Schreiber, G. Mutational analysis of the IFNAR1 binding site on IFNα2 reveals the architecture of a weak ligand-receptor binding-site. J. Mol. Biol. 353, 271–281 (2005)
Ferguson, B. J. et al. Biophysical and cell-based evidence for differential interactions between the death domains of CD95/Fas and FADD. Cell Death Differ. 14, 1717–1719 (2007)
Itoh, N. & Nagata, S. A novel protein domain required for apoptosis. Mutational analysis of human Fas antigen. J. Biol. Chem. 268, 10932–10937 (1993)
Kim, H. E., Du, F., Fang, M. & Wang, X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl Acad. Sci. USA 102, 17545–17550 (2005)
Boatright, K. M. et al. A unified model for apical caspase activation. Mol. Cell 11, 529–541 (2003)
Acknowledgements
We thank S. Snipas for protein sequencing and technical assistance, J. Reed for providing Fas cDNA and A. Bobkov for the AUC. This work was supported by a P30 CA030199 cancer center grant and R01AA017238 to S.J.R.; PO1CA69381 to G.S.S.; and MCEXT-033534 to R.S. Data measured at beamline X29 of the National Synchrotron Light Source were also supported by Biological and Environmental Research DOE, and National Center for Research Resources NIH. Earlier stages of the work were supported by a LLS scholarship to S.J.R. S.J.R. is currently a V Foundation scholar.
Author Contributions F.L.S. performed and evaluated in vivo studies. J.J.L. and M.K.D. performed cloning, protein expression and crystallization. C.P. performed biochemical analyses. H.R., B.S. and in particular R.S. performed data collection and structure solution. E.M. performed EM-studies. G.S.S. participated in study design and evaluation. All authors discussed the work. S.J.R. participated in and oversaw all aspects of the work and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Atomic coordinates and experimental structure factors have been deposited within the Protein Data Bank and are accessible under the code 3EZQ.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-5, a Supplementary Discussion, Supplementary Tables 1-2, Supplementary Methods and Supplementary References (PDF 1797 kb)
Rights and permissions
About this article
Cite this article
Scott, F., Stec, B., Pop, C. et al. The Fas–FADD death domain complex structure unravels signalling by receptor clustering. Nature 457, 1019–1022 (2009). https://doi.org/10.1038/nature07606
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07606
This article is cited by
-
Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation
Cell Research (2023)
-
Regulated cell death pathways and their roles in homeostasis, infection, inflammation, and tumorigenesis
Experimental & Molecular Medicine (2023)
-
Identification of biomarkers for glycaemic deterioration in type 2 diabetes
Nature Communications (2023)
-
Therapeutic approaches targeting CD95L/CD95 signaling in cancer and autoimmune diseases
Cell Death & Disease (2022)
-
FAS receptor regulates NOTCH activity through ERK-JAG1 axis activation and controls oral cancer stemness ability and pulmonary metastasis
Cell Death Discovery (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.