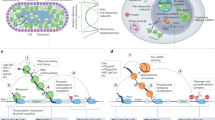
Abstract
Ligand binding to structural elements in the non-coding regions of messenger RNA modulates gene expression1,2. Ligands such as free metabolites or other small molecules directly bind and induce conformational changes in regulatory RNA elements known as riboswitches1,2,3,4. Other types of RNA switches are activated by complexed metabolites—for example, RNA-ligated metabolites such as aminoacyl-charged transfer RNA in the T-box system5, or protein-bound metabolites in the glucose- or amino-acid-stimulated terminator-anti-terminator systems6,7. All of these switch types are found in bacteria, fungi and plants8,9,10. Here we report an RNA switch in human vascular endothelial growth factor-A (VEGFA, also known as VEGF) mRNA 3′ untranslated region (UTR) that integrates signals from interferon (IFN)-γ and hypoxia to regulate VEGFA translation in myeloid cells. Analogous to riboswitches, the VEGFA 3′ UTR undergoes a binary conformational change in response to environmental signals. However, the VEGFA 3′ UTR switch is metabolite-independent, and the conformational change is dictated by mutually exclusive, stimulus-dependent binding of proteins, namely, the IFN-γ-activated inhibitor of translation complex11,12 and heterogeneous nuclear ribonucleoprotein L (HNRNPL, also known as hnRNP L). We speculate that the VEGFA switch represents the founding member of a family of signal-mediated, protein-dependent RNA switches that evolved to regulate gene expression in multicellular animals in which the precise integration of disparate inputs may be more important than the rapidity of response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mandal, M. & Breaker, R. R. Gene regulation by riboswitches. Nature Rev. Mol. Cell Biol. 5, 451–463 (2004)
Grundy, F. J. & Henkin, T. M. Regulation of gene expression by effectors that bind to RNA. Curr. Opin. Microbiol. 7, 126–131 (2004)
Winkler, W., Nahvi, A. & Breaker, R. R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002)
Cromie, M. J., Shi, Y., Latifi, T. & Groisman, E. A. An RNA sensor for intracellular Mg2+ . Cell 125, 71–84 (2006)
Grundy, F. J. & Henkin, T. M. tRNA as a positive regulator of transcription antitermination in B. subtilis . Cell 74, 475–482 (1993)
Merino, E., Babitzke, P. & Yanofsky, C. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol. 177, 6362–6370 (1995)
Schilling, O., Langbein, I., Muller, M., Schmalisch, M. H. & Stulke, J. A protein-dependent riboswitch controlling ptsGHI operon expression in Bacillus subtilis: RNA structure rather than sequence provides interaction specificity. Nucleic Acids Res. 32, 2853–2864 (2004)
Batey, R. T. Structures of regulatory elements in mRNAs. Curr. Opin. Struct. Biol. 16, 299–306 (2006)
Cheah, M. T., Wachter, A., Sudarsan, N. & Breaker, R. R. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 447, 497–500 (2007)
Wachter, A. et al. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19, 3437–3450 (2007)
Mazumder, B. et al. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115, 187–198 (2003)
Sampath, P. et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell 119, 195–208 (2004)
Braunstein, S. et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell 28, 501–512 (2007)
Bastide, A. et al. An upstream open reading frame within an IRES controls expression of a specific VEGF-A isoform. Nucleic Acids Res. 36, 2434–2445 (2008)
Ferrara, N. & Davis-Smyth, T. The biology of vascular endothelial growth factor. Endocr. Rev. 18, 4–25 (1997)
Ray, P. S. & Fox, P. L. A post-transcriptional pathway represses monocyte VEGF-A expression and angiogenic activity. EMBO J. 26, 3360–3372 (2007)
Mukhopadhyay, R. et al. DAPK-ZIPK-L13a axis constitutes a negative-feedback module regulating inflammatory gene expression. Mol. Cell 32, 371–382 (2008)
Liu, L. et al. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell 21, 521–531 (2006)
Sampath, P., Mazumder, B., Seshadri, V. & Fox, P. L. Transcript-selective translational silencing by gamma interferon is directed by a novel structural element in the ceruloplasmin mRNA 3′ untranslated region. Mol. Cell. Biol. 23, 1509–1519 (2003)
Piñol-Roma, S., Swanson, M. S., Gall, J. G. & Dreyfuss, G. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J. Cell Biol. 109, 2575–2587 (1989)
Claffey, K. P. et al. Identification of a human VPF/VEGF 3′ untranslated region mediating hypoxia-induced mRNA stability. Mol. Biol. Cell 9, 469–481 (1998)
Shih, S. C. & Claffey, K. P. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J. Biol. Chem. 274, 1359–1365 (1999)
Jia, J., Arif, A., Ray, P. S. & Fox, P. L. WHEP domains direct noncanonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Mol. Cell 29, 679–690 (2008)
Jaeger, J. A., Turner, D. H. & Zuker, M. Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA 86, 7706–7710 (1989)
Knapp, G. Enzymatic approaches to probing RNA secondary and tertiary structure. Methods Enzymol. 180, 192–212 (1989)
Brion, P. & Westhof, E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 26, 113–137 (1997)
Huthoff, H. & Berkhout, B. Two alternating structures of the HIV-1 leader RNA. RNA 7, 143–157 (2001)
Sudarsan, N. et al. Tandem riboswitch architectures exhibit complex gene control functions. Science 314, 300–304 (2006)
Yaman, I. et al. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113, 519–531 (2003)
Merrick, W. C. & Hensold, J. O. Analysis of eukaryotic translation in purified and semipurified systems. Curr. Protocols. Cell Biol. Chapter 11, Unit–11.9 (2001)
Mazumder, B., Seshadri, V., Imataka, H., Sonenberg, N. & Fox, P. L. Translational silencing of ceruloplasmin requires the essential elements of mRNA circularization: Poly(A) tail, poly(A)-binding protein, and eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 21, 6440–6449 (2001)
Acknowledgements
We are grateful to D. Driscoll and T. M. Henkin for helpful discussions. This work was supported by National Institutes of Health grants P01 HL29582, R01 HL67725 and P01 HL76491 (to P.L.F.), and R01 DK60596 (to M.H.).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-12 with Legends (PDF 1994 kb)
Rights and permissions
About this article
Cite this article
Ray, P., Jia, J., Yao, P. et al. A stress-responsive RNA switch regulates VEGFA expression. Nature 457, 915–919 (2009). https://doi.org/10.1038/nature07598
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07598
This article is cited by
-
Probing the dynamic RNA structurome and its functions
Nature Reviews Genetics (2023)
-
Deciphering the role of RNA structure in translation efficiency
BMC Bioinformatics (2022)
-
The energy-spectrum of bicompatible sequences
Algorithms for Molecular Biology (2021)
-
FBXO16-mediated hnRNPL ubiquitination and degradation plays a tumor suppressor role in ovarian cancer
Cell Death & Disease (2021)
-
Mex-3B induces apoptosis by inhibiting miR-92a access to the Bim-3′UTR
Oncogene (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.