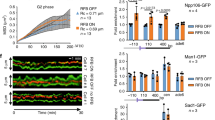
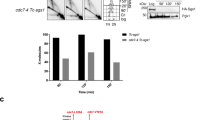
Abstract
Replication by template switch is thought to mediate DNA damage-bypass and fillings of gaps. Gap-filling repair requires homologous recombination as well as Rad18- and Rad5-mediated proliferating cell nuclear antigen (PCNA) polyubiquitylation. However, it is unclear whether these processes are coordinated, and the physical evidence for Rad18–Rad5-dependent template switch at replication forks is still elusive. Here we show, using genetic and physical approaches, that in budding yeast (Saccharomyces cerevisiae) Rad18 is required for the formation of X-shaped sister chromatid junctions (SCJs) at damaged replication forks through a process involving PCNA polyubiquitylation and the ubiquitin-conjugating enzymes Mms2 and Ubc13. The Rad18–Mms2-mediated damage-bypass through SCJs requires the small ubiquitin-like modifier (SUMO)-conjugating enzyme Ubc9 and SUMOylated PCNA, and is coordinated with Rad51-dependent recombination events. We propose that the Rad18–Rad5–Mms2-dependent SCJs represent template switch events. Altogether, our results unmask a role for PCNA ubiquitylation and SUMOylation pathways in promoting transient damage-induced replication-coupled recombination events involving sister chromatids at replication forks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Prakash, L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 184, 471–478 (1981)
Lopes, M., Foiani, M. & Sogo, J. M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21, 15–27 (2006)
Branzei, D. & Foiani, M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst.) 6, 994–1003 (2007)
Lehmann, A. R. & Fuchs, R. P. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst.) 5, 1495–1498 (2006)
Fabre, F., Chan, A., Heyer, W. D. & Gangloff, S. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA 99, 16887–16892 (2002)
Symington, L. S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630–670 (2002)
San Filippo, J., Sung, P. & Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008)
Gangavarapu, V., Prakash, S. & Prakash, L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae . Mol. Cell. Biol. 27, 7758–7764 (2007)
Zhang, H. & Lawrence, C. W. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl Acad. Sci. USA 102, 15954–15959 (2005)
Torres-Ramos, C. A., Prakash, S. & Prakash, L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae . Mol. Cell. Biol. 22, 2419–2426 (2002)
Haracska, L. et al. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae . Mol. Cell. Biol. 24, 4267–4274 (2004)
Higgins, N. P., Kato, K. & Strauss, B. A model for replication repair in mammalian cells. J. Mol. Biol. 101, 417–425 (1976)
Goldfless, S. J. et al. DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol. Cell 21, 595–604 (2006)
Branzei, D. & Foiani, M. Template switching: from replication fork repair to genome rearrangements. Cell 131, 1228–1230 (2007)
Jentsch, S., McGrath, J. P. & Varshavsky, A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329, 131–134 (1987)
Hofmann, R. M. & Pickart, C. M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 (1999)
Ulrich, H. D. & Jentsch, S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19, 3388–3397 (2000)
Bailly, V. et al. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8, 811–820 (1994)
Branzei, D. & Foiani, M. Regulation of DNA repair throughout the cell cycle. Nature Rev. Mol. Cell Biol. 4, 297–308 (2008)
Hoege, C. et al. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002)
Stelter, P. & Ulrich, H. D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188–191 (2003)
Kannouche, P. L., Wing, J. & Lehmann, A. R. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, 491–500 (2004)
Geiss-Friedlander, R. & Melchior, F. Concepts in sumoylation: a decade on. Nature Rev. Mol. Cell Biol. 8, 947–956 (2007)
Pfander, B. et al. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 (2005)
Papouli, E. et al. Crosstalk between SUMO and Ubiquitin on PCNA Is Mediated by Recruitment of the Helicase Srs2p. Mol. Cell 19, 123–133 (2005)
Lawrence, C. W. & Christensen, R. B. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 139, 866–876 (1979)
Aboussekhra, A. et al. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 17, 7211–7219 (1989)
Schiestl, R. H., Prakash, S. & Prakash, L. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124, 817–831 (1990)
Palladino, F. & Klein, H. L. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics 132, 23–37 (1992)
Krejci, L. et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 (2003)
Veaute, X. et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 (2003)
Branzei, D. et al. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127, 509–522 (2006)
Liberi, G. et al. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 19, 339–350 (2005)
Mankouri, H. W., Ngo, H. P. & Hickson, I. D. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell 18, 4062–4073 (2007)
Wu, L. & Hickson, I. D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 (2003)
Johnson, E. S. Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 (2004)
Onoda, F., Seki, M., Miyajima, A. & Enomoto, T. Involvement of SGS1 in DNA damage-induced heteroallelic recombination that requires RAD52 in Saccharomyces cerevisiae . Mol. Gen. Genet. 264, 702–708 (2001)
Branzei, D., Seki, M. & Enomoto, T. Rad18/Rad5/Mms2-mediated polyubiquitination of PCNA is implicated in replication completion during replication stress. Genes Cells 9, 1031–1042 (2004)
Tateishi, S. et al. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl Acad. Sci. USA 97, 7927–7932 (2000)
Zhao, G. Y. et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell 25, 663–675 (2007)
Motegi, A. et al. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J. Cell Biol. 175, 703–708 (2006)
Motegi, A. et al. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae . Mol. Cell. Biol. 26, 1424–1433 (2006)
Daee, D. L., Mertz, T. & Lahue, R. S. Postreplication repair inhibits CAG·CTG repeat expansions in Saccharomyces cerevisiae . Mol. Cell. Biol. 27, 102–110 (2007)
Johnson, R. E. et al. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 12, 3807–3818 (1992)
Sung, P. & Klein, H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nature Rev. Mol. Cell Biol. 7, 739–750 (2006)
Otsuki, M. et al. Functional interactions between BLM and XRCC3 in the cell. J. Cell Biol. 179, 53–63 (2007)
Szuts, D. et al. Role for RAD18 in homologous recombination in DT40 cells. Mol. Cell. Biol. 26, 8032–8041 (2006)
Tateishi, S. et al. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 23, 474–481 (2003)
Yamashita, Y. M. et al. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 21, 5558–5566 (2002)
Shekhar, M. P. et al. Rad6 overexpression induces multinucleation, centrosome amplification, abnormal mitosis, aneuploidy, and transformation. Cancer Res. 62, 2115–2124 (2002)
Acknowledgements
We thank S. Jentsch and H. Ulrich for yeast strains, T. Enomoto for critical reading, and all members of our laboratories for helpful discussions. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro to D.B., Association for International Cancer Research and European Community GENICA grant to M.F. and D.B., and partly by European Community DNA Repair grant, Telethon, MIUR, and Ministry of Health to M.F. D.B. was partly supported by the Buzzati-Traverso foundation.
Author Contributions D.B. conceived the project, designed and performed the experiments, and wrote the paper. F.V. quantified the 2D gels, performed data analysis and provided technical help. M.F. discussed the results, analysed the data, edited the manuscript and provided scientific advice and financial support.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-12 with Legends, Supplementary Table 1 and Supplementary References. (PDF 2065 kb)
Rights and permissions
About this article
Cite this article
Branzei, D., Vanoli, F. & Foiani, M. SUMOylation regulates Rad18-mediated template switch. Nature 456, 915–920 (2008). https://doi.org/10.1038/nature07587
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07587
This article is cited by
-
The COMPASS subunit Spp1 protects nascent DNA at the Tus/Ter replication fork barrier by limiting DNA availability to nucleases
Nature Communications (2023)
-
Break-induced replication orchestrates resection-dependent template switching
Nature (2023)
-
Mechanism for inverted-repeat recombination induced by a replication fork barrier
Nature Communications (2022)
-
Rad51-mediated replication of damaged templates relies on monoSUMOylated DDK kinase
Nature Communications (2022)
-
Participation of the HIM1 gene of yeast Saccharomyces cerevisiae in the error-free branch of post-replicative repair and role Polη in him1-dependent mutagenesis
Current Genetics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.