Abstract
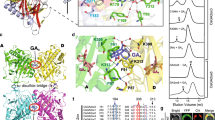
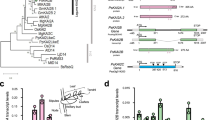
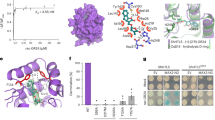
Gibberellins (GAs) are phytohormones essential for many developmental processes in plants1. A nuclear GA receptor, GIBBERELLIN INSENSITIVE DWARF1 (GID1), has a primary structure similar to that of the hormone-sensitive lipases (HSLs)2,3. Here we analyse the crystal structure of Oryza sativa GID1 (OsGID1) bound with GA4 and GA3 at 1.9 Å resolution. The overall structure of both complexes shows an α/β-hydrolase fold similar to that of HSLs except for an amino-terminal lid. The GA-binding pocket corresponds to the substrate-binding site of HSLs. On the basis of the OsGID1 structure, we mutagenized important residues for GA binding and examined their binding activities. Almost all of them showed very little or no activity, confirming that the residues revealed by structural analysis are important for GA binding. The replacement of Ile 133 with Leu or Val—residues corresponding to those of the lycophyte Selaginella moellendorffii GID1s—caused an increase in the binding affinity for GA34, a 2β-hydroxylated GA4. These observations indicate that GID1 originated from HSL and was further modified to have higher affinity and more strict selectivity for bioactive GAs by adapting the amino acids involved in GA binding in the course of plant evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Olszewski, N., Sun, T. P. & Gubler, F. Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14 (Suppl.). S61–S80 (2002)
Ueguchi-Tanaka, M. et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698 (2005)
Ueguchi-Tanaka, M. et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19, 2140–2155 (2007)
Davies, P. J. (ed.) Plant Hormones: Biosynthesis, Signal Transduction, Action! 3rd edn (Kluwer, 2004)
Ueguchi-Tanaka, M., Nakajima, M., Ashikari, M. & Matsuoka, M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58, 183–198 (2007)
Itoh, H., Ueguchi-Tanaka, M. & Matsuoka, M. Molecular biology of gibberellins signaling in higher plants. Int. Rev. Cell Mol. Biol. 268, 191–221 (2008)
Hirano, K. et al. The GID1-mediated gibberellin perception mechanism is conserved in the Lycophyte Selaginella moellendorffii but not in the Bryophyte Physcomitrella patens . Plant Cell 19, 3058–3079 (2007)
Yasumura, Y., Crumpton-Taylor, M., Fuentes, S. & Harberd, N. P. Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land–plant evolution. Curr. Biol. 17, 1225–1230 (2007)
Yeaman, S. J. Hormone-sensitive lipase—new roles for an old enzyme. Biochem. J. 379, 11–22 (2004)
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 (2004)
Ileperuma, N. R. et al. High-resolution crystal structure of plant carboxylesterase AeCXE1, from Actinidia eriantha, and its complex with a high-affinity inhibitor paraoxon. Biochemistry 46, 1851–1859 (2007)
Ollis, D. L. et al. The α/β hydrolase fold. Protein Eng. 5, 197–211 (1992)
Holm, C., Davis, R. C., Osterlund, T., Schotz, M. C. & Fredrikson, G. Identification of the active site serine of hormone-sensitive lipase by site-directed mutagenesis. FEBS Lett. 344, 234–238 (1994)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276, 307–326 (1997)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Perrakis, A., Morris, R. & Lamzin, V. S. Automated protein model building combined with iterative structure refinement. Nature Struct. Biol. 6, 458–463 (1999)
Roussel, A. & Cambillau, C. Silicon Graphics Geometry Partners Directory, Vol. 81 77–78 (Silicon Graphics, Mountain View, 1991)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Acknowledgements
We thank M. Kawamura, M. Hattori, Y. Yamamoto, K. Aya and T. Matsubara for technical assistance; R. L. Ordonio and M. Tanrikulu for editing of this manuscript; and T. Shimizu, RIKEN, and N. Shimizu, JASRI at SPring-8, for assistance with data collection. This project was funded by the Target Protein Research Program (M.M. and H.K.), Scientific Research (M.M., and M.U.-T.), and Special Coordination Funds for Promoting Science and Technology (H.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by research fellowships from the Japan Society for the Promotion of Science (A.S.).
Author Contributions M.M. and H.K. conceived and designed the project; A.S. performed construct design, purification, crystallization and structure determinations; Y.N. assisted purification, crystallization and heavy-atom derivative preparation; T.N. solved and refined the structures; M.U.-T. conducted experimental work including cloning, mutation, expression, purification and two-hybrid assays; H.O. assisted purification; M.N. performed binding assays and helped with critical discussions of the work; M.U.-T., H.K. and M.M. wrote the manuscript; and A.S. and T.N. edited the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Table 1, Supplementary Figures 1-9 and Supplementary References. (PDF 4325 kb)
Rights and permissions
About this article
Cite this article
Shimada, A., Ueguchi-Tanaka, M., Nakatsu, T. et al. Structural basis for gibberellin recognition by its receptor GID1. Nature 456, 520–523 (2008). https://doi.org/10.1038/nature07546
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07546
This article is cited by
-
An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity
Nature Chemical Biology (2024)
-
The master growth regulator DELLA binding to histone H2A is essential for DELLA-mediated global transcription regulation
Nature Plants (2023)
-
Identification of a new gibberellin receptor agonist, diphegaractin, by a cell-free chemical screening system
Communications Biology (2023)
-
Analysis of the Physiological Roles and Mode of Actions of Phthalimides as GA Signal Regulator in Rice
Journal of Plant Growth Regulation (2023)
-
Strigolactones: diversity, perception, and hydrolysis
Phytochemistry Reviews (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.