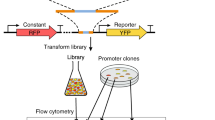
Abstract
Transcription factor binding sites are being discovered at a rapid pace1,2. It is now necessary to turn attention towards understanding how these sites work in combination to influence gene expression. Quantitative models that accurately predict gene expression from promoter sequence3,4,5 will be a crucial part of solving this problem. Here we present such a model, based on the analysis of synthetic promoter libraries in yeast (Saccharomyces cerevisiae). Thermodynamic models based only on the equilibrium binding of transcription factors to DNA and to each other captured a large fraction of the variation in expression in every library. Thermodynamic analysis of these libraries uncovered several phenomena in our system, including cooperativity and the effects of weak binding sites. When applied to the S. cerevisiae genome, a model of repression by Mig1 (which was trained on synthetic promoters) predicts a number of Mig1-regulated genes that lack significant Mig1-binding sites in their promoters. The success of the thermodynamic approach suggests that the information encoded by combinations of cis-regulatory sites is interpreted primarily through simple protein–DNA and protein–protein interactions, with complicated biochemical reactions—such as nucleosome modifications—being downstream events. Quantitative analyses of synthetic promoter libraries will be an important tool in unravelling the rules underlying combinatorial cis-regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Harbison, C. T. et al. Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104 (2004)
Hu, Z., Killion, P. J. & Iyer, V. R. Genetic reconstruction of a functional transcriptional regulatory network. Nature Genet. 39, 683–687 (2007)
Beer, M. A. & Tavazoie, S. Predicting gene expression from sequence. Cell 117, 185–198 (2004)
Bussemaker, H. J., Li, H. & Siggia, E. D. Regulatory element detection using correlation with expression. Nature Genet. 27, 167–171 (2001)
Das, D., Banerjee, N. & Zhang, M. Q. Interacting models of cooperative gene regulation. Proc. Natl Acad. Sci. USA 101, 16234–16239 (2004)
Segal, E., Raveh-Sadka, T., Schroeder, M., Unnerstall, U. & Gaul, U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451, 535–540 (2008)
Zinzen, R. P., Senger, K., Levine, M. & Papatsenko, D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr. Biol. 16, 1358–1365 (2006)
Murphy, K. F., Balazsi, G. & Collins, J. J. Combinatorial promoter design for engineering noisy gene expression. Proc. Natl Acad. Sci. USA 104, 12726–12731 (2007)
Ligr, M., Siddharthan, R., Cross, F. R. & Siggia, E. D. Gene expression from random libraries of yeast promoters. Genetics 172, 2113–2122 (2006)
Cox, R. S., Surette, M. G. & Elowitz, M. B. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 3, 145 (2007)
Shea, M. A. & Ackers, G. K. The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J. Mol. Biol. 181, 211–230 (1985)
Buchler, N. E., Gerland, U. & Hwa, T. On schemes of combinatorial transcription logic. Proc. Natl Acad. Sci. USA 100, 5136–5141 (2003)
Ptashne, M. & Gann, A. Genes and Signals (Cold Spring Harbor Laboratory Press, 2002)
Tanay, A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 16, 962–972 (2006)
Lutfiyya, L. L. et al. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae . Genetics 150, 1377–1391 (1998)
Hertz, G. Z. & Stormo, G. D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15, 563–577 (1999)
Matys, V. et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31, 374–378 (2003)
Nehlin, J. O. & Ronne, H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 9, 2891–2898 (1990)
Monteiro, P. T. et al. YEASTRACT-DISCOVERER: new tools to improve the analysis of transcriptional regulatory associations in Saccharomyces cerevisiae . Nucleic Acids Res. 36, D132–D136 (2008)
Teixeira, M. C. et al. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae . Nucleic Acids Res. 34, D446–D451 (2006)
Ptashne, M. & Gann, A. Transcriptional activation by recruitment. Nature 386, 569–577 (1997)
Gietz, R. D. & Schiestl, R. H. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2, 38–41 (2007)
Brachmann, C. B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998)
Kaniak, A., Xue, Z., Macool, D., Kim, J. H. & Johnston, M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae . Eukaryot. Cell 3, 221–231 (2004)
Gish, W. WU BLAST. 〈http://blast.wustl.edu〉 (1995-, 2008)
Cliften, P. et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301, 71–76 (2003)
Hertz, G. Z. & Stormo, G. D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15, 563–577 (1999)
Nehlin, J. O. & Ronne, H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 9, 2891–2898 (1990)
Acknowledgements
We thank R. Mitra, G. Stormo, M. Johnston, K. Varley, S. Doniger and members of the Cohen laboratory for discussions and suggestions, and J. Sabina for technical help with LacZ experiments. B.A.C. and J.G. were supported by the NIH (R01 GM078222) and E.D.S. was supported by the NSF (DMR0129848). J.G. was also supported by an NSF Graduate Research Fellowship (DGE-0202737).
Author Contributions J.G. performed all experiments and analyses. B.A.C. and J.G. designed the experiments and wrote the paper. E.D.S. conceived the idea of applying the thermodynamic model to the synthetic promoter libraries, and contributed to its development.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures S1-S4 and Supplementary Tables S1-S9. (PDF 6838 kb)
Rights and permissions
About this article
Cite this article
Gertz, J., Siggia, E. & Cohen, B. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457, 215–218 (2009). https://doi.org/10.1038/nature07521
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07521
This article is cited by
-
Chance promoter activities illuminate the origins of eukaryotic intergenic transcriptions
Nature Communications (2023)
-
Gene regulation in Escherichia coli is commonly selected for both high plasticity and low noise
Nature Ecology & Evolution (2022)
-
The metabolic enzyme hexokinase 2 localizes to the nucleus in AML and normal haematopoietic stem and progenitor cells to maintain stemness
Nature Cell Biology (2022)
-
Quantitative-enhancer-FACS-seq (QeFS) reveals epistatic interactions among motifs within transcriptional enhancers in developing Drosophila tissue
Genome Biology (2021)
-
Deciphering eukaryotic gene-regulatory logic with 100 million random promoters
Nature Biotechnology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.