Abstract
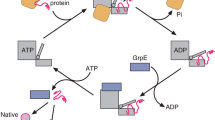
A subset of essential cellular proteins requires the assistance of chaperonins (in Escherichia coli, GroEL and GroES), double-ring complexes in which the two rings act alternately to bind, encapsulate and fold a wide range of nascent or stress-denatured proteins1,2,3,4,5. This process starts by the trapping of a substrate protein on hydrophobic surfaces in the central cavity of a GroEL ring6,7,8,9,10. Then, binding of ATP and co-chaperonin GroES to that ring ejects the non-native protein from its binding sites, through forced unfolding or other major conformational changes, and encloses it in a hydrophilic chamber for folding11,12,13,14,15. ATP hydrolysis and subsequent ATP binding to the opposite ring trigger dissociation of the chamber and release of the substrate protein3. The bacteriophage T4 requires its own version of GroES, gp31, which forms a taller folding chamber, to fold the major viral capsid protein gp23 (refs 16–20). Polypeptides are known to fold inside the chaperonin complex, but the conformation of an encapsulated protein has not previously been visualized. Here we present structures of gp23–chaperonin complexes, showing both the initial captured state and the final, close-to-native state with gp23 encapsulated in the folding chamber. Although the chamber is expanded, it is still barely large enough to contain the elongated gp23 monomer, explaining why the GroEL–GroES complex is not able to fold gp23 and showing how the chaperonin structure distorts to enclose a large, physiological substrate protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
The three-dimensional reconstructions for the two binary complexes and the three ternary complexes are deposited in the EBI-MSD EMD database with accession codes EMD-1544 through to EMD-1548.
References
Houry, W. A. et al. Identification of in vivo substrates of the chaperonin GroEL. Nature 402, 147–154 (1999)
Kerner, M. J. et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli . Cell 122, 209–220 (2005)
Rye, H. S. et al. GroEL-GroES cycling: ATP and nonnative polypeptide direct alternation of folding-active rings. Cell 97, 325–338 (1999)
Sigler, P. B. et al. Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem. 67, 581–608 (1998)
Horwich, A. L., Fenton, W. A., Chapman, E. & Farr, G. W. Two families of chaperonin: Physiology and mechanism. Annu. Rev. Cell Dev. Biol. 23, 115–145 (2007)
Braig, K. et al. The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature 371, 578–586 (1994)
Fenton, W. A., Kashi, Y., Furtak, K. & Horwich, A. L. Residues in chaperonin GroEL required for polypeptide binding and release. Nature 371, 614–619 (1994)
Lin, Z., Schwartz, F. P. & Eisenstein, E. The hydrophobic nature of GroEL-substrate binding. J. Biol. Chem. 270, 1011–1014 (1995)
Farr, G. W. et al. Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell 100, 561–573 (2000)
Elad, N. et al. Topologies of a substrate protein bound to the chaperonin GroEL. Mol. Cell 26, 415–426 (2007)
Xu, Z., Horwich, A. L. & Sigler, P. B. The crystal structure of the asymmetric GroEL–GroES–(ADP)7 chaperonin complex. Nature 388, 741–750 (1997)
Motojima, F. et al. Substrate polypeptide presents a load on the apical domains of the chaperonin GroEL. Proc. Natl Acad. Sci. USA 101, 15005–15012 (2004)
Lin, Z. & Rye, H. S. Expansion and compression of a protein folding intermediate by GroEL. Mol. Cell 16, 23–34 (2004)
Lin, Z., Madan, D. & Rye, H. S. GroEL stimulates protein folding through forced unfolding. Nature Struct. Mol. Biol. 15, 303–311 (2008)
Mayhew, M. et al. Protein folding in the central cavity of the GroEL–GroES chaperonin complex. Nature 379, 420–426 (1996)
Laemmli, U. K., Beguin, F. & Gujer-Kellenberger, G. A factor preventing the major head protein of bacteriophage T4 from random aggregation. J. Mol. Biol. 47, 69–85 (1970)
van der Vies, S. M., Gatenby, A. A. & Georgopoulos, C. Bacteriophage T4 encodes a co-chaperonin that can substitute for Escherichia coli GroES in protein folding. Nature 368, 654–656 (1994)
Hunt, J. F., van der Vies, S. M., Henry, L. & Deisenhofer, J. Structural adaptations in the specialized bacteriophage T4 co-chaperonin Gp31 expand the size of the Anfinsen cage. Cell 90, 361–371 (1997)
Bakkes, P. J., Faber, B. W., van Heerikhuizen, H. & van der Vies, S. M. The T4-encoded cochaperonin, gp31, has unique properties that explain its requirement for the folding of the T4 major capsid protein. Proc. Natl Acad. Sci. USA 102, 8144–8149 (2005)
Clare, D. K. et al. An expanded protein folding cage in the GroEL-gp31 complex. J. Mol. Biol. 358, 905–911 (2006)
Elad, N., Clare, D. K., Saibil, H. R. & Orlova, E. V. Detection and separation of heterogeneity in molecular complexes by statistical analysis of their two-dimensional projections. J. Struct. Biol. 162, 108–120 (2008)
van Duijn, E. et al. Monitoring macromolecular complexes involved in the chaperonin-assisted protein folding cycle by mass spectrometry. Nature Methods 2, 371–376 (2005)
van Duijn, E., Heck, A. J. & van der Vies, S. M. Inter-ring communication allows the GroEL chaperonin complex to distinguish between different substrates. Protein Sci. 16, 956–965 (2007)
Ranson, N. A. et al. Allosteric signaling of ATP hydrolysis in GroEL-GroES complexes. Nature Struct. Mol. Biol. 13, 147–152 (2006)
Fokine, A. et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl Acad. Sci. USA 102, 7163–7168 (2005)
van Duijn, E. et al. Tandem mass spectrometry of intact GroEL-substrate complexes reveals substrate-specific conformational changes in the trans ring. J. Am. Chem. Soc. 128, 4694–4702 (2006)
Snyder, L. & Tarkowski, H. J. The N terminus of the head protein of T4 bacteriophage directs proteins to the GroEL chaperonin. J. Mol. Biol. 345, 375–386 (2005)
Falke, S. et al. The 13 angstroms structure of a chaperonin GroEL-protein substrate complex by cryo-electron microscopy. J. Mol. Biol. 348, 219–230 (2005)
Tang, Y. C. et al. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125, 903–914 (2006)
Machida, K. et al. Hydrophilic residues 526KNDAAD531 in the flexible C-terminal region of the chaperonin GroEL are critical for substrate protein folding within the central cavity. J. Biol. Chem. 283, 6886–6896 (2008)
Chaudhry, C. et al. Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: Function, structure and energetics. EMBO J. 22, 4877–4887 (2003)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Crowther, R. A., Henderson, R. & Smith, J. M. MRC image processing programs. J. Struct. Biol. 116, 9–16 (1996)
Frank, J. et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
van Heel, M. et al. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996)
Navaza, J. et al. On the fitting of model electron densities into EM reconstructions: A reciprocal-space formulation. Acta Crystallogr. D 58, 1820–1825 (2002)
Acknowledgements
We thank R. Westlake, D. Houldershaw and L. Wang for computing and EM support. This work was carried out at the School of Crystallography, Birkbeck College, and was supported by a Wellcome Trust programme grant, EU 3D EM Network of Excellence and 3D Repertoire grants.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-3 with Legends, Supplementary Table 1, Supplementary Notes and Discussion and Supplementary References (PDF 1537 kb)
Rights and permissions
About this article
Cite this article
Clare, D., Bakkes, P., van Heerikhuizen, H. et al. Chaperonin complex with a newly folded protein encapsulated in the folding chamber. Nature 457, 107–110 (2009). https://doi.org/10.1038/nature07479
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07479
This article is cited by
-
Molecular chaperones and their denaturing effect on client proteins
Journal of Biomolecular NMR (2021)
-
Distinct Stabilities of the Structurally Homologous Heptameric Co-Chaperonins GroES and gp31
Journal of the American Society for Mass Spectrometry (2019)
-
Blob-ology and biology of cryo-EM: an interview with Helen Saibil
BMC Biology (2017)
-
Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nature Communications (2017)
-
Molecular chaperones: providing a safe place to weather a midlife protein-folding crisis
Nature Structural & Molecular Biology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.