Abstract
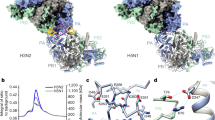
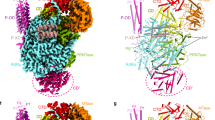
Influenza A virus is a major human and animal pathogen with the potential to cause catastrophic loss of life. The virus reproduces rapidly, mutates frequently and occasionally crosses species barriers. The recent emergence in Asia of avian influenza related to highly pathogenic forms of the human virus has highlighted the urgent need for new effective treatments1. Here we demonstrate the importance to viral replication of a subunit interface in the viral RNA polymerase, thereby providing a new set of potential drug binding sites entirely independent of surface antigen type. No current medication targets this heterotrimeric polymerase complex. All three subunits, PB1, PB2 and PA, are required for both transcription and replication2,3,4. PB1 carries the polymerase active site, PB2 includes the capped-RNA recognition domain, and PA is involved in assembly of the functional complex5,6,7, but so far very little structural information has been reported for any of them8,9,10,11. We describe the crystal structure of a large fragment of one subunit (PA) of influenza A RNA polymerase bound to a fragment of another subunit (PB1). The carboxy-terminal domain of PA forms a novel fold, and forms a deep, highly hydrophobic groove into which the amino-terminal residues of PB1 can fit by forming a 310 helix.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Peiris, J. S., de Jong, M. D. & Guan, Y. Avian influenza virus (H5N1): a threat to human health. Clin. Microbiol. Rev. 20, 243–267 (2007)
Horisberger, M. A. The large P proteins of influenza A viruses are composed of one acidic and two basic polypeptides. Virology 107, 302–305 (1980)
Braam, J., Ulmanen, I. & Krug, R. M. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell 34, 611–618 (1983)
Nagata, K., Kawaguchi, A. & Naito, T. Host factors for replication and transcription of the influenza virus genome. Rev. Med. Virol. 18, 247–260 (2008)
Fodor, E., Mingay, L. J., Crow, M., Deng, T. & Brownlee, G. G. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase promotes the generation of defective interfering RNAs. J. Virol. 77, 5017–5020 (2003)
Kawaguchi, A., Naito, T. & Nagata, K. Involvement of influenza virus PA subunit in assembly of functional RNA polymerase complexes. J. Virol. 79, 732–744 (2005)
Deng, T., Sharps, J. L. & Brownlee, G. G. Role of the influenza virus heterotrimeric RNA polymerase complex in the initiation of replication. J. Gen. Virol. 87, 3373–3377 (2006)
Area, E. et al. 3D structure of the influenza virus polymerase complex: localization of subunit domains. Proc. Natl Acad. Sci. USA 101, 308–313 (2004)
Torreira, E. et al. Three-dimensional model for the isolated recombinant influenza virus polymerase heterotrimer. Nucleic Acids Res. 35, 3774–3783 (2007)
Tarendeau, F. et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nature Struct. Mol. Biol. 14, 229–233 (2007)
Guilligay, D. et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nature Struct. Mol. Biol. 15, 500–506 (2008)
Elton, D., Digard, P., Tiley, L. & Ortin, J. in Current Topics in Influenza Virology (ed. Kawaoka, Y.) 1–92 (Horizon Scientific Press, 2005)
Plotch, S. J., Bouloy, M., Ulmanen, I. & Krug, R. M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23, 847–858 (1981)
Taubenberger, J. K. et al. Characterization of the 1918 influenza virus polymerase genes. Nature 437, 889–893 (2005)
Zurcher, T., de la Luna, S., Sanz-Ezquerro, J. J., Nieto, A. & Ortin, J. Mutational analysis of the influenza virus A/Victoria/3/75 PA protein: studies of interaction with PB1 protein and identification of a dominant negative mutant. J. Gen. Virol. 77, 1745–1749 (1996)
Perez, D. R. & Donis, R. O. Functional analysis of PA binding by influenza A virus PB1: effects on polymerase activity and viral infectivity. J. Virol. 75, 8127–8136 (2001)
Ohtsu, Y., Honda, Y., Sakata, Y., Kato, H. & Toyoda, T. Fine mapping of the subunit binding sites of influenza virus RNA polymerase. Microbiol. Immunol. 46, 167–175 (2002)
Ghanem, A. et al. Peptide-mediated interference with influenza A virus polymerase. J. Virol. 81, 7801–7804 (2007)
Hara, K., Schmidt, F. I., Crow, M. & Brownlee, G. G. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 80, 7789–7798 (2006)
Fodor, E. et al. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76, 8989–9001 (2002)
Huarte, M. et al. Threonine 157 of influenza virus PA polymerase subunit modulates RNA replication in infectious viruses. J. Virol. 77, 6007–6013 (2003)
Regan, J. F., Liang, Y. & Parslow, T. G. Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J. Virol. 80, 252–261 (2006)
Hsieh, H. P. & Hsu, J. T. Strategies of development of antiviral agents directed against influenza virus replication. Curr. Pharm. Des. 13, 3531–3542 (2007)
World Health Organization A revision of the system of nomenclature for influenza viruses: a WHO memorandum. Bull. World Health Organ. 58, 585–591 (1980)
Kim, C. U. et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: Design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc. 119, 681–690 (1997)
von Itzstein, M. et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363, 418–423 (1993)
Russell, R. J. et al. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443, 45–49 (2006)
Liu, Y., Zhang, J. & Xu, W. Recent progress in rational drug design of neuraminidase inhibitors. Curr. Med. Chem. 14, 2872–2891 (2007)
Wang, C., Takeuchi, K., Pinto, L. H. & Lamb, R. A. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 67, 5585–5594 (1993)
Stouffer, A. L. et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature 451, 596–599 (2008)
Kawaguchi, A. & Nagata, K. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J. 26, 4566–4575 (2007)
Turan, K. et al. Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic Acids Res. 32, 643–652 (2004)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
Terwilliger, T. C. SOLVE and RESOLVE: Automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Roussel, A. & Cambillau, C. in Silicon Graphics Geometry Partner Directory 77–78 (Silicon Graphics, 1989)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Laskowski, R. A., Moss, D. S. & Thornton, J. M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231, 1049–1067 (1993)
Acknowledgements
We thank the staff at the SPring-8 BL41XU and Photon Factory BL5A beamlines for assistance with data collection, and M. Yao and Y. Zhou for help with data processing. This work was supported in part by the ISS applied research partnership program (S.-Y.P.).
Author Contributions E.O., J.R.H.T., K.N. and S.-Y.P. conceived and designed the project. E.O. carried out complex purification and crystallization, and pull-down assays. E.O., H.Y., F.K., N.S. and S.-Y.P. conducted experimental work including cloning, expression, purification, binding assays, data collection and structure analysis. A.K. and K.N. tested the transcriptional activity of mutants. E.O., J.R.H.T. and S.-Y.P. wrote the manuscript, and all authors discussed the results and conclusions.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information 1
The file contains Supplementary Methods and additional references, Supplementary Figures 1-2 with Legends and Supplementary Table 1. (PDF 2809 kb)
Rights and permissions
About this article
Cite this article
Obayashi, E., Yoshida, H., Kawai, F. et al. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454, 1127–1131 (2008). https://doi.org/10.1038/nature07225
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07225
This article is cited by
-
Upregulation of galectin-3 in influenza A virus infection promotes viral RNA synthesis through its association with viral PA protein
Journal of Biomedical Science (2023)
-
The ubiquitination landscape of the influenza A virus polymerase
Nature Communications (2023)
-
An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates
Virus Genes (2022)
-
Evolution of KIPPIS as a versatile platform for evaluating intracellularly functional peptide aptamers
Scientific Reports (2021)
-
Therapeutic p28 peptide targets essential H1N1 influenza virus proteins: insights from docking and molecular dynamics simulations
Molecular Diversity (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.