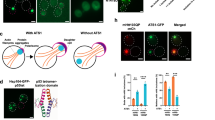
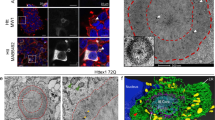
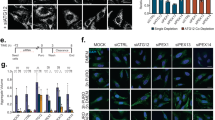
Abstract
The accumulation of misfolded proteins in intracellular amyloid inclusions, typical of many neurodegenerative disorders including Huntington’s and prion disease, is thought to occur after failure of the cellular protein quality control mechanisms. Here we examine the formation of misfolded protein inclusions in the eukaryotic cytosol of yeast and mammalian cell culture models. We identify two intracellular compartments for the sequestration of misfolded cytosolic proteins. Partition of quality control substrates to either compartment seems to depend on their ubiquitination status and aggregation state. Soluble ubiquitinated misfolded proteins accumulate in a juxtanuclear compartment where proteasomes are concentrated. In contrast, terminally aggregated proteins are sequestered in a perivacuolar inclusion. Notably, disease-associated Huntingtin and prion proteins are preferentially directed to the perivacuolar compartment. Enhancing ubiquitination of a prion protein suffices to promote its delivery to the juxtanuclear inclusion. Our findings provide a framework for understanding the preferential accumulation of amyloidogenic proteins in inclusions linked to human disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bence, N. F., Sampat, R. M. & Kopito, R. R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292, 1552–1555 (2001)
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006)
Gidalevitz, T., Ben-Zvi, A., Ho, K. H., Brignull, H. R. & Morimoto, R. I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474 (2006)
Muchowski, P. J. & Wacker, J. L. Modulation of neurodegeneration by molecular chaperones. Nature Rev. Neurosci. 6, 11–22 (2005)
Outeiro, T. F. & Lindquist, S. Yeast cells provide insight into α-synuclein biology and pathobiology. Science 302, 1772–1775 (2003)
Schaffar, G. et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol. Cell 15, 95–105 (2004)
Lesne, S. et al. A specific amyloid-β protein assembly in the brain impairs memory. Nature 440, 352–357 (2006)
Rubinsztein, D. C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 (2006)
Iwata, A. et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl Acad. Sci. USA 102, 13135–13140 (2005)
Taylor, J. P. et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 12, 749–757 (2003)
Rideout, H. J., Lang-Rollin, I. & Stefanis, L. Involvement of macroautophagy in the dissolution of neuronal inclusions. Int. J. Biochem. Cell Biol. 36, 2551–2562 (2004)
Yorimitsu, T. & Klionsky, D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12 (Suppl 2). 1542–1552 (2005)
Matsumoto, G., Kim, S. & Morimoto, R. I. Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. J. Biol. Chem. 281, 4477–4485 (2006)
Bucciantini, M. et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 (2002)
Sherman, M. Y. & Goldberg, A. L. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron 29, 15–32 (2001)
Matsumoto, G., Stojanovic, A., Holmberg, C. I., Kim, S. & Morimoto, R. I. Structural properties and neuronal toxicity of amyotrophic lateral sclerosis-associated Cu/Zn superoxide dismutase 1 aggregates. J. Cell Biol. 171, 75–85 (2005)
Krobitsch, S. & Lindquist, S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl Acad. Sci. USA 97, 1589–1594 (2000)
Kamhi-Nesher, S. et al. A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell 12, 1711–1723 (2001)
Huyer, G. et al. A striking quality control subcompartment in Saccharomyces cerevisiae: the endoplasmic reticulum-associated compartment. Mol. Biol. Cell 15, 908–921 (2004)
Kopito, R. R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 (2000)
Kruse, K. B., Brodsky, J. L. & McCracken, A. A. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human α-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol. Biol. Cell 17, 203–212 (2006)
McClellan, A. J., Tam, S., Kaganovich, D. & Frydman, J. Protein quality control: chaperones culling corrupt conformations. Nature Cell Biol. 7, 736–741 (2005)
Betting, J. & Seufert, W. A yeast Ubc9 mutant protein with temperature-sensitive in vivo function is subject to conditional proteolysis by a ubiquitin- and proteasome-dependent pathway. J. Biol. Chem. 271, 25790–25796 (1996)
Tongaonkar, P., Beck, K., Shinde, U. P. & Madura, K. Characterization of a temperature-sensitive mutant of a ubiquitin-conjugating enzyme and its use as a heat-inducible degradation signal. Anal. Biochem. 272, 263–269 (1999)
McClellan, A. J., Scott, M. D. & Frydman, J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121, 739–748 (2005)
Vang, S. et al. Actin mutations in hypertrophic and dilated cardiomyopathy cause inefficient protein folding and perturbed filament formation. FEBS J. 272, 2037–2049 (2005)
Feldman, D. E., Thulasiraman, V., Ferreyra, R. G. & Frydman, J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol. Cell 4, 1051–1061 (1999)
Mateus, C. & Avery, S. V. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 16, 1313–1323 (2000)
Duennwald, M. L., Jagadish, S., Giorgini, F., Muchowski, P. J. & Lindquist, S. A network of protein interactions determines polyglutamine toxicity. Proc. Natl Acad. Sci. USA 103, 11051–11056 (2006)
Lippincott-Schwartz, J. & Patterson, G. H. Development and use of fluorescent protein markers in living cells. Science 300, 87–91 (2003)
Rujano, M. A. et al. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 4, e417 (2006)
Erjavec, N. & Nystrom, T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae . Proc. Natl Acad. Sci. USA 104, 10877–10881 (2007)
Enenkel, C., Lehmann, A. & Kloetzel, P. M. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 17, 6144–6154 (1998)
Tkach, J. M. & Glover, J. R. Amino acid substitutions in the C-terminal AAA+ module of Hsp104 prevent substrate recognition by disrupting oligomerization and cause high temperature inactivation. J. Biol. Chem. 279, 35692–35701 (2004)
Sarkar, S. et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nature Chem. Biol. 3, 331–338 (2007)
Szeto, J. et al. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy 2, 189–199 (2006)
Kuma, A., Matsui, M. & Mizushima, N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 3, 323–328 (2007)
Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007)
Swaminathan, S., Amerik, A. Y. & Hochstrasser, M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10, 2583–2594 (1999)
Balch, W. E., Morimoto, R. I., Dillin, A. & Kelly, J. W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008)
Siegel, S. J., Bieschke, J., Powers, E. T. & Kelly, J. W. The oxidative stress metabolite 4-hydroxynonenal promotes Alzheimer protofibril formation. Biochemistry 46, 1503–1510 (2007)
Cohen, E., Bieschke, J., Perciavalle, R. M., Kelly, J. W. & Dillin, A. Opposing activities protect against age-onset proteotoxicity. Science 313, 1604–1610 (2006)
Webb, J. L., Ravikumar, B., Atkins, J., Skepper, J. N. & Rubinsztein, D. C. α-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 278, 25009–25013 (2003)
Kruse, K. B., Brodsky, J. L. & McCracken, A. A. Autophagy: an ER protein quality control process. Autophagy 2, 135–137 (2006)
Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004)
Barral, J. M., Broadley, S. A., Schaffar, G. & Hartl, F. U. Roles of molecular chaperones in protein misfolding diseases. Semin. Cell Dev. Biol. 15, 17–29 (2004)
Tam, S., Geller, R., Spiess, C. & Frydman, J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nature Cell Biol. 8, 1155–1162 (2006)
Melville, M. W., McClellan, A. J., Meyer, A. S., Darveau, A. & Frydman, J. The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol. Cell. Biol. 23, 3141–3151 (2003)
Adams, A., Gottschling, D., Kaiser, C. & Stearns, T. Methods in Yeast Genetics (Cold Spring Harbor Laboratory Press, 1997)
Shaner, N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnol. 22, 1567–1572 (2004)
Prinz, A., Hartmann, E. & Kalies, K. U. Sec61p is the main ribosome receptor in the endoplasmic reticulum of Saccharomyces cerevisiae . Biol. Chem. 381, 1025–1029 (2000)
Ghislain, M., Udvardy, A. & Mann, C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366, 358–362 (1993)
Chen, P., Johnson, P., Sommer, T., Jentsch, S. & Hochstrasser, M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT α 2 repressor. Cell 74, 357–369 (1993)
Winzeler, E. A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999)
Mulholland, J. et al. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 125, 381–391 (1994)
Wright, R. & Rine, J. Transmission electron microscopy and immunocytochemical studies of yeast: analysis of HMG-CoA reductase overproduction by electron microscopy. Methods in cell biology 31, 473–512 (1989)
Acknowledgements
We thank R. Tsien, S. Michaelis, J. Glover, C. Enenkel and V. Albanese for reagents; J. Mulholland for electron microscopy and deconvolution microscopy help; S. Yamada and W. J. Nelson for help with the live cell microscopy. We thank R. Andino, W. Burkholder, J. England, R. Geller, M. Kaganovich, E. Bennett, J. Nelson and members of the Frydman laboratory for discussions and comments on the manuscript. This work was supported by grants from NIH to R.K. and J.F.
Author Contributions J.F. directed the project; D.K. and J.F. designed and interpreted all experiments; R.K. participated in the initial fluorescence microscopy experiments; D.K. performed all experiments. D.K. and J.F. wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
The file contains Supplementary Figures S1-S8 with Legends and Legends to Supplementary Movies 1-2. (PDF 1023 kb)
Supplementary Movie 1
The file contains Supplementary Movie 1. (MPG 1779 kb)
Supplementary Movie 2
The file contains Supplementary Movie 2. (MPG 1796 kb)
Rights and permissions
About this article
Cite this article
Kaganovich, D., Kopito, R. & Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 (2008). https://doi.org/10.1038/nature07195
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07195
This article is cited by
-
Calcineurin stimulation by Cnb1p overproduction mitigates protein aggregation and α-synuclein toxicity in a yeast model of synucleinopathy
Cell Communication and Signaling (2023)
-
The GET pathway is a major bottleneck for maintaining proteostasis in Saccharomyces cerevisiae
Scientific Reports (2023)
-
The yeast guanine nucleotide exchange factor Sec7 is a bottleneck in spatial protein quality control and detoxifies neurological disease proteins
Scientific Reports (2023)
-
Nuclear and cytoplasmic spatial protein quality control is coordinated by nuclear–vacuolar junctions and perinuclear ESCRT
Nature Cell Biology (2023)
-
Artificial Hsp104-mediated systems for re-localizing protein aggregates
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.