Abstract
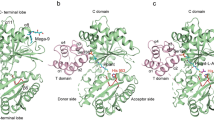
Non-ribosomal peptide synthetases (NRPS) and polyketide synthases (PKS) found in bacteria, fungi and plants use two different types of thioesterases for the production of highly active biological compounds1,2. Type I thioesterases (TEI) catalyse the release step from the assembly line3 of the final product where it is transported from one reaction centre to the next as a thioester linked to a 4′-phosphopantetheine (4′-PP) cofactor that is covalently attached to thiolation (T) domains4,5,6,7,8,9. The second enzyme involved in the synthesis of these secondary metabolites, the type II thioesterase (TEII), is a crucial repair enzyme for the regeneration of functional 4′-PP cofactors of holo-T domains of NRPS and PKS systems10,11,12. Mispriming of 4′-PP cofactors by acetyl- and short-chain acyl-residues interrupts the biosynthetic system. This repair reaction is very important, because roughly 80% of CoA, the precursor of the 4′-PP cofactor, is acetylated in bacteria13. Here we report the three-dimensional structure of a type II thioesterase from Bacillus subtilis free and in complex with a T domain. Comparison with structures of TEI enzymes3,14 shows the basis for substrate selectivity and the different modes of interaction of TEII and TEI enzymes with T domains. Furthermore, we show that the TEII enzyme exists in several conformations of which only one is selected on interaction with its native substrate, a modified holo-T domain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Walsh, C. T. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science 303, 1805–1810 (2004)
Kopp, F. & Marahiel, M. A. Macrocyclization strategies in polyketide and nonribosomal peptide biosynthesis. Nat. Prod. Rep. 24, 735–749 (2007).
Bruner, S. D. et al. Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE. Structure 10, 301–310 (2002)
Kim, Y. & Prestegard, J. H. A dynamic model for the structure of acyl carrier protein in solution. Biochemistry 28, 8792–8797 (1989)
Koglin, A. et al. Conformational switches modulate protein interactions in peptide antibiotic synthetases. Science 312, 273–276 (2006)
Leibundgut, M., Jenni, S., Frick, C. & Ban, N. Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science 316, 288–290 (2007)
Findlow, S. C., Winsor, C., Simpson, T. J., Crosby, J. & Crump, M. P. Solution structure and dynamics of oxytetracycline polyketide synthase acyl carrier protein from Streptomyces rimosus . Biochemistry 42, 8423–8433 (2003)
Johnson, M. A., Peti, W., Herrmann, T., Wilson, I. A. & Wüthrich, K. Solution structure of Asl1650, an acyl carrier protein from Anabaena sp. PCC 7120 with a variant phosphopantetheinylation-site sequence. Protein Sci. 15, 1030–1041 (2006)
Zornetzer, G. A., Fox, B. G. & Markley, J. L. Solution structures of spinach acyl carrier protein with decanoate and stearate. Biochemistry 45, 5217–5227 (2006)
Schwarzer, D., Mootz, H. D., Linne, U. & Marahiel, M. A. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl Acad. Sci. USA 99, 14083–14088 (2002).
Linne, U., Schwarzer, D., Schroeder, G. N. & Marahiel, M. A. Mutational analysis of a type II thioesterase associated with nonribosomal peptide synthesis. Eur. J. Biochem. 271, 1536–1545 (2004).
Yeh, E., Kohli, R. M., Bruner, S. D. & Walsh, C. T. Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-S-enzyme that cannot be elongated. ChemBioChem 5, 1290–1293 (2004).
Conti, E., Stachelhaus, T., Marahiel, M. A. & Brick, P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16, 4174–4183 (1997)
Gehring, A. M., Mori, I. & Walsh, C. T. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37, 2648–2659 (1998)
Steller, S. et al. Initiation of surfactin biosynthesis and the role of the SrfD-thioesterase protein. Biochemistry 43, 11331–11343 (2004)
Samel, S. A., Schoenafinger, G., Knappe, T. A., Marahiel, M. A. & Essen, L.-O. Structural and functional insights into a peptide bond-forming bidomain from a nonribosomal peptide synthetase. Structure 15, 781–792 (2007).
Schneider, A. & Marahiel, M. A. Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in Bacillus subtilis . Arch. Microbiol. 169, 404–410 (1998)
Zuiderweg, E. R. P. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry 41, 1–7 (2002)
Frueh, D. P. et al. Dynamic thiolation–thioesterase structure of a non-ribosomal peptide synthetase. Nature 10.1038/nature07162 (this issue)
Gardner, K. H. & Kay, L. E. Production and incorporation of 15N, 13C, 2H (1H-δ1 methyl) isoleucine into proteins for multidimensional NMR studies. J. Am. Chem. Soc. 119, 7599–7600 (1997)
Frueh, D. P., Vosburg, D. A., Walsh, C. T. & Wagner, G. Determination of all NOes in 1H- 13C-Me-ILV-U-2H-15N proteins with two time-shared experiments. J. Biomol. NMR 34, 31–40 (2006).
Lai, J. R., Koglin, A. & Walsh, C. T. Carrier protein structure and recognition in polyketide and nonribosomal peptide biosynthesis. Biochemistry 45, 14869–14879 (2006).
Schwarzer, D., Mootz, H. D., Linne, U. & Marahiel, M. A. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl Acad. Sci. USA 99, 14083–14088 (2002)
Jenni, S. et al. Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science 316, 254–261 (2007)
Pervushin, K., Riek, R., Wider, G. & Wüthrich, K. Attenuated T 2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl Acad. Sci. USA 94, 12366–12371 (1997)
Sattler, M., Schleucher, J. & Griesinger, C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 (1999)
Ferentz, A. E. & Wagner, G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q. Rev. Biophys. 33, 29–65 (2000)
Güntert, P., Mumenthaler, C. & Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 (1997)
Nederveen, A. J. et al. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59, 662–672 (2005)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Ikura, M., Kay, L. E. & Bax, A. Improved three-dimensional 1H-13C-1H correlation spectroscopy of a 13C-labeled protein using constant-time evolution. J. Biomol. NMR 1, 299–304 (1991)
Salzmann, M., Pervushin, K., Wider, G., Senn, H. & Wüthrich, K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc. Natl Acad. Sci. USA 95, 13585–13590 (1998)
Bax, A., Clore, M. & Gronenborn, A. M. 1H-1H Correlation via isotropic mixing of 13C magentization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 88, 425–431 (1990)
Güntert, P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 (2004)
Cornilescu, G., Delaglio, F. & Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999)
Liang, J., Edelsbrunner, H. & Woodward, C. Anatomy of protein pockets and cavities: Measurement of binding site geometry and implications for ligand design. Protein Sci. 7, 1884–1897 (1998)
Dundas, J. et al. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated resiudes. Nucleic Acids Res. 34, W116–W118 (2006)
Cobas, C. F. & Sardina, J. Nuclear magnetic resonance data processing. MestRe-C: A software package for desktop computers. Concepts Magn. Reson. A 19, 80–96 (2003)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Belshaw, P. J., Walsh, C. T. & Stachelhaus, T. Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis. Science 284, 486–489 (1999)
Sieber, S. A., Walsh, C. T. & Marahiel, M. A. Loading peptidyl-coenzyme A onto peptidyl carrier proteins: a novel approach in characterizing macrocyclization by thioesterase domains. J. Am. Chem. Soc. 125, 10862–10866 (2003)
Meier, J. L., Mercer, A. C., Rivera, H. & Burkart, M. D. Synthesis and evaluation of bioorthogonal pantetheine analogues for in vivo protein modification. J. Am. Chem. Soc. 128, 12174–12184 (2006)
Acknowledgements
We thank M. Strieker for editing the manuscript and B. Schaefer for her help and support in sample preparation. We thank Chi Scientific Inc. for the fast and high quality supply of the substrate peptides. The research was funded by the research grant BE-19/11 of the Deutsche Forschungsgemeinschaft (F.B. and M.A.M.); an enclosed fellowship (A.K.); the Centre for Biomolecular Magnetic Resonance at the University Frankfurt; the Cluster of Excellence Frankfurt (Macromolecular Complexes); and the Volkswagen Foundation (P.G.). A.K. thanks the Human Frontier Science Program Organization for a long-term fellowship awarded in April 2007.
Author Contributions A.K., V.D., F.B. and M.A.M. designed the experiments and defined the research for the SrfTEII. A.K., V.D. and C.T.W. wrote the manuscript and designed the experiments for the complex structure. A.K., F.B. and F.L. conducted the research including protein expression, data acquisition, resonance assignment and structure calculation. V.R.R. helped with resonance assignment and structure calculation of the SrfTEII. P.G. provided the volumes of the cavities of SrfTEI and of the complex SrfTEII–TycC3 T domain and supported the complex structure calculation. D.P.F. and G.W. recorded the NMR spectra of the protein complex. A.K. calculated the structure of the isolated SrfTEII, the structure of the protein complex (SrfTEII and TycC3–PCP) and performed all titration experiments. M.A.M. and M.R.M. provided the expression vectors for SrfTEII and the TycC3 T domain. M.A.M. provided the purification protocol and biochemical characterization of the SrfTEII. E.R.S. provided the non-hydrolysable CoA derivate and synthesized the peptide and amino acid modified substrate analogues.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary information
The file contains Supplementary Notes, Supplementary Figures 1-8 with Legends, Supplementary Tables 1-2 and additional references. (PDF 2115 kb)
Rights and permissions
About this article
Cite this article
Koglin, A., Löhr, F., Bernhard, F. et al. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature 454, 907–911 (2008). https://doi.org/10.1038/nature07161
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07161
This article is cited by
-
Research advances in the identification of regulatory mechanisms of surfactin production by Bacillus: a review
Microbial Cell Factories (2024)
-
Integrated Biofilm Modification and Transcriptional Analysis for Improving Fengycin Production in Bacillus amyloliquefaciens
Probiotics and Antimicrobial Proteins (2024)
-
Efficient determination of the accessible conformation space of multi-domain complexes based on EPR PELDOR data
Journal of Biomolecular NMR (2023)
-
Genomics-driven discovery of a biosynthetic gene cluster required for the synthesis of BII-Rafflesfungin from the fungus Phoma sp. F3723
BMC Genomics (2019)
-
Characterization and heterologous expression of the neoabyssomicin/abyssomicin biosynthetic gene cluster from Streptomyces koyangensis SCSIO 5802
Microbial Cell Factories (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.