Abstract
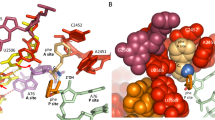
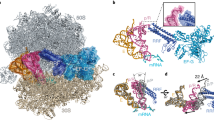
At termination of protein synthesis, type I release factors promote hydrolysis of the peptidyl-transfer RNA linkage in response to recognition of a stop codon. Here we describe the crystal structure of the Thermus thermophilus 70S ribosome in complex with the release factor RF1, tRNA and a messenger RNA containing a UAA stop codon, at 3.2 Å resolution. The stop codon is recognized in a pocket formed by conserved elements of RF1, including its PxT recognition motif, and 16S ribosomal RNA. The codon and the 30S subunit A site undergo an induced fit that results in stabilization of a conformation of RF1 that promotes its interaction with the peptidyl transferase centre. Unexpectedly, the main-chain amide group of Gln 230 in the universally conserved GGQ motif of the factor is positioned to contribute directly to peptidyl-tRNA hydrolysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Capecchi, M. R. Polypeptide chain termination in vitro: isolation of a release factor. Proc. Natl Acad. Sci. USA 58, 1144–1151 (1967)
Caskey, C. T., Beaudet, A. L., Scolnick, E. M. & Rosman, M. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc. Natl Acad. Sci. USA 68, 3163–3167 (1971)
Vogel, Z., Zamir, A. & Elson, D. The possible involvement of peptidyl transferase in the termination step of protein biosynthesis. Biochemistry 8, 5161–5168 (1969)
Scolnick, E., Tompkins, R., Caskey, T. & Nirenberg, M. Release factors differing in specificity for terminator codons. Proc. Natl Acad. Sci. USA 61, 768–774 (1968)
Capecchi, M. R. & Klein, H. A. Characterization of three proteins involved in polypeptide chain termination. Cold Spring Harb. Symp. Quant. Biol. 34, 469–477 (1969)
Scolnick, E. M. & Caskey, C. T. Peptide chain termination. V. The role of release factors in mRNA terminator codon recognition. Proc. Natl Acad. Sci. USA 64, 1235–1241 (1969)
Freistroffer, D. V., Kwiatkowski, M., Buckingham, R. H. & Ehrenberg, M. The accuracy of codon recognition by polypeptide release factors. Proc. Natl Acad. Sci. USA 97, 2046–2051 (2000)
Petry, S. et al. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123, 1255–1266 (2005)
Klaholz, B. P. et al. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 421, 90–94 (2003)
Rawat, U. B. et al. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421, 87–90 (2003)
Ito, K., Uno, M. & Nakamura, Y. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403, 680–684 (2000)
Nakamura, Y. & Ito, K. A tripeptide discriminator for stop codon recognition. FEBS Lett. 514, 30–33 (2002)
Ogle, J. M., Carter, A. P. & Ramakrishnan, V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28, 259–266 (2003)
Ogle, J. M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001)
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006)
Youngman, E. M., Cochella, L., Brunelle, J. L., He, S. & Green, R. Two distinct conformations of the conserved RNA-rich decoding center of the small ribosomal subunit are recognized by tRNAs and release factors. Cold Spring Harb. Symp. Quant. Biol. 71, 545–549 (2006)
Youngman, E. M., He, S. L., Nikstad, L. J. & Green, R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol. Cell 28, 533–543 (2007)
Frolova, L. Y. et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5, 1014–1020 (1999)
Seit-Nebi, A., Frolova, L., Justesen, J. & Kisselev, L. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 29, 3982–3987 (2001)
Mora, L. et al. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol. Microbiol. 47, 267–275 (2003)
Shaw, J. J. & Green, R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol. Cell 28, 458–467 (2007)
Trobro, S. & Aqvist, J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol. Cell 27, 758–766 (2007)
Song, H. et al. The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100, 311–321 (2000)
Seit Nebi, A., Frolova, L., Ivanova, N., Poltaraus, A. & Kisselev, L. Mutation of a glutamine residue in the universal tripeptide GGQ in human eRF1 termination factor does not cause complete loss of its activity. Mol. Biol. (Mosk.) 34, 899–900 (2000)
Schmeing, T. M., Huang, K. S., Strobel, S. A. & Steitz, T. A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438, 520–524 (2005)
Vestergaard, B. et al. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell 8, 1375–1382 (2001)
Schuwirth, B. S. et al. Structures of the bacterial ribosome at 3.5 A resolution. Science 310, 827–834 (2005)
Amort, M. et al. An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination. Nucleic Acids Res. 35, 5130–5140 (2007)
Polacek, N. et al. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol. Cell 11, 103–112 (2003)
Youngman, E. M., Brunelle, J. L., Kochaniak, A. B. & Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117, 589–599 (2004)
Maegley, K. A., Admiraal, S. J. & Herschlag, D. Ras-catalyzed hydrolysis of GTP: A new perspective from model studies. Proc. Natl Acad. Sci. USA 93, 8160–8166 (1996)
Li, G. & Zhang, X. C. GTP hydrolysis mechanism of Ras-like GTPases. J. Mol. Biol. 340, 921–932 (2004)
Schmeing, T. M., Huang, K. S., Kitchen, D. E., Strobel, S. A. & Steitz, T. A. Structural insights into the roles of water and the 2' hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell 20, 437–448 (2005)
Schmeing, T. M., Moore, P. B. & Steitz, T. A. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 9, 1345–1352 (2003)
Zavialov, A. V., Mora, L., Buckingham, R. H. & Ehrenberg, M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell 10, 789–798 (2002)
Dincbas-Renqvist, V. et al. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 19, 6900–6907 (2000)
Shin, D. H. et al. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J. Mol. Biol. 341, 227–239 (2004)
Zoldak, G. et al. Release factors 2 from Escherichia coli and Thermus thermophilus: structural, spectroscopic and microcalorimetric studies. Nucleic Acids Res. 35, 1343–1353 (2007)
Vestergaard, B. et al. The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Mol. Cell 20, 929–938 (2005)
Ali, I. K., Lancaster, L., Feinberg, J., Joseph, S. & Noller, H. F. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell 23, 865–874 (2006)
Brunger, A. T. et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Yusupov, M. M. et al. Crystal structure of the ribosome at 5.5 A resolution. Science 292, 883–896 (2001)
Acknowledgements
We thank B. Scott for his input through many stages of this work, and D. Ermolenko, L. Horan, L. Lancaster, B. Scott and D. Staple for comments on the manuscript. We are grateful to the beamline staff at SSRL, ALS and APS for their support during screening and data collection, and to Crystal Chan for her help in using the Berkeley Fermentation Facility. This work was supported by grants from the NIH and NSF (to H.F.N.) and by a fellowship from the Danish Research Council (to M.L.).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Table S1, Supplementary Figures S1-S11 and References. (PDF 1769 kb)
Rights and permissions
About this article
Cite this article
Laurberg, M., Asahara, H., Korostelev, A. et al. Structural basis for translation termination on the 70S ribosome. Nature 454, 852–857 (2008). https://doi.org/10.1038/nature07115
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07115
This article is cited by
-
The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center
Nature Chemical Biology (2024)
-
Antibiotic thermorubin tethers ribosomal subunits and impedes A-site interactions to perturb protein synthesis in bacteria
Nature Communications (2023)
-
Structural basis for translation inhibition by the glycosylated drosocin peptide
Nature Chemical Biology (2023)
-
Human mtRF1 terminates COX1 translation and its ablation induces mitochondrial ribosome-associated quality control
Nature Communications (2022)
-
Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.