Abstract
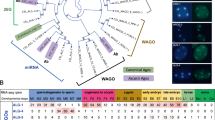
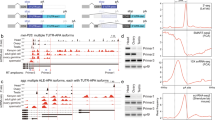
Drosophila endogenous small RNAs are categorized according to their mechanisms of biogenesis and the Argonaute protein to which they bind. MicroRNAs are a class of ubiquitously expressed RNAs of ∼22 nucleotides in length, which arise from structured precursors through the action of Drosha–Pasha and Dicer-1–Loquacious complexes1,2,3,4,5,6,7. These join Argonaute-1 to regulate gene expression8,9. A second endogenous small RNA class, the Piwi-interacting RNAs, bind Piwi proteins and suppress transposons10,11. Piwi-interacting RNAs are restricted to the gonad, and at least a subset of these arises by Piwi-catalysed cleavage of single-stranded RNAs12,13. Here we show that Drosophila generates a third small RNA class, endogenous small interfering RNAs, in both gonadal and somatic tissues. Production of these RNAs requires Dicer-2, but a subset depends preferentially on Loquacious1,4,5 rather than the canonical Dicer-2 partner, R2D2 (ref. 14). Endogenous small interfering RNAs arise both from convergent transcription units and from structured genomic loci in a tissue-specific fashion. They predominantly join Argonaute-2 and have the capacity, as a class, to target both protein-coding genes and mobile elements. These observations expand the repertoire of small RNAs in Drosophila, adding a class that blurs distinctions based on known biogenesis mechanisms and functional roles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
Small RNA sequences were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE11086.
References
Forstemann, K. et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3, e236 (2005)
Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003)
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F. & Hannon, G. J. Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235 (2004)
Saito, K., Ishizuka, A., Siomi, H. & Siomi, M. C. Processing of pre-microRNAs by the Dicer-1–Loquacious complex in Drosophila cells. PLoS Biol. 3, e235 (2005)
Jiang, F. et al. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila . Genes Dev. 19, 1674–1679 (2005)
Lee, Y. S. et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81 (2004)
Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001)
Eulalio, A., Huntzinger, E. & Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 132, 9–14 (2008)
Bushati, N. & Cohen, S. M. microRNA functions. Annu. Rev. Cell Dev. Biol. 23, 175–205 (2007)
Aravin, A. A., Hannon, G. J. & Brennecke, J. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761–764 (2007)
Klattenhoff, C. & Theurkauf, W. Biogenesis and germline functions of piRNAs. Development 135, 3–9 (2008)
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila . Cell 128, 1089–1103 (2007)
Gunawardane, L. S. et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila . Science 315, 1587–1590 (2007)
Liu, Q. et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301, 1921–1925 (2003)
Ruby, J. G. et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17, 1850–1864 (2007)
Galiana-Arnoux, D., Dostert, C., Schneemann, A., Hoffmann, J. A. & Imler, J. L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila . Nature Immunol. 7, 590–597 (2006)
Wang, X. H. et al. RNA interference directs innate immunity against viruses in adult Drosophila . Science 312, 452–454 (2006)
Saito, K. et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222 (2006)
Vagin, V. V. et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 (2006)
Rehwinkel, J. et al. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster . Mol. Cell. Biol. 26, 2965–2975 (2006)
Allen, E., Xie, Z., Gustafson, A. M. & Carrington, J. C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 (2005)
Tomari, Y., Du, T. & Zamore, P. D. Sorting of Drosophila small silencing RNAs. Cell 130, 299–308 (2007)
Forstemann, K., Horwich, M. D., Wee, L., Tomari, Y. & Zamore, P. D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell 130, 287–297 (2007)
Ruby, J. G. et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans . Cell 127, 1193–1207 (2006)
Sijen, T., Steiner, F. A., Thijssen, K. L. & Plasterk, R. H. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247 (2007)
Pak, J. & Fire, A. Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science 315, 241–244 (2007)
Yigit, E. et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747–757 (2006)
Tam, O. H. et al. Pseudogene-derived siRNAs regulate gene expression in mouse oocytes. Nature advance online publication, 10.1038/nature06904 (10 April 2008)
Watanabe, T. et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature advance online publication, 10.1038/nature06908 (10 April 2008)
Okamura, K., Ishizuka, A., Siomi, H. & Siomi, M. C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18, 1655–1666 (2004)
Stark, A. et al. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 17, 1865–1879 (2007)
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005)
Acknowledgements
We thank R. Carthew, H. Siomi, P. Zamore and D. Smith for reagents. We are grateful to M. Rooks, E. Hodges and D. McCombie for help with deep sequencing. B.C. was supported by the German Academic Exchange Service. C.D.M. is a Beckman fellow of the Watson School of Biological Sciences and is supported by an NSF Graduate Research Fellowship. R.Z. is a Special Fellow of the Leukemia and Lymphoma Society. M.D. is an Engelhorn fellow of the Watson School of Biological Sciences. J.B. is supported by the Ernst Schering foundation. A.S. is supported by an HFSP fellowship. This work was supported in part from grants from the NIH to G.J.H. and N.P. and a gift from K. W. Davis (G.J.H.).
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
The file contains Supplementary Methods, Supplementary Tables S1-S2, Supplementary Figures S1-S13 and additional references. (PDF 1479 kb)
Rights and permissions
About this article
Cite this article
Czech, B., Malone, C., Zhou, R. et al. An endogenous small interfering RNA pathway in Drosophila. Nature 453, 798–802 (2008). https://doi.org/10.1038/nature07007
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07007
This article is cited by
-
Modeling early germline immunization after horizontal transfer of transposable elements reveals internal piRNA cluster heterogeneity
BMC Biology (2023)
-
Small RNAs, spermatogenesis, and male infertility: a decade of retrospect
Reproductive Biology and Endocrinology (2023)
-
Mitochondrially mediated RNA interference, a retrograde signaling system affecting nuclear gene expression
Heredity (2023)
-
Discovery of the major 15–30 nt mammalian small RNAs, their biogenesis and function
Nature Communications (2023)
-
Structure of the Dicer-2–R2D2 heterodimer bound to a small RNA duplex
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.