Abstract
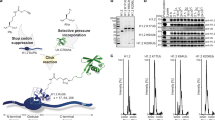
Numerous post-translational modifications of histones have been described in organisms ranging from yeast to humans1. Growing evidence for dynamic regulation of these modifications, position- and modification-specific protein interactions, and biochemical crosstalk between modifications has strengthened the ‘histone code’ hypothesis, in which histone modifications are integral to choreographing the expression of the genome1,2. One such modification, ubiquitylation of histone H2B (uH2B) on lysine 120 (K120) in humans3, and lysine 123 in yeast4, has been correlated with enhanced methylation of lysine 79 (K79) of histone H3 (refs 5–8), by K79-specific methyltransferase Dot1 (KMT4)9,10,11. However, the specific function of uH2B in this crosstalk pathway is not understood. Here we demonstrate, using chemically ubiquitylated H2B, a direct stimulation of hDot1L-mediated intranucleosomal methylation of H3 K79. Two traceless orthogonal expressed protein ligation (EPL) reactions were used to ubiquitylate H2B site-specifically. This strategy, using a photolytic ligation auxiliary and a desulphurization reaction, should be generally applicable to the chemical ubiquitylation of other proteins. Reconstitution of our uH2B into chemically defined nucleosomes, followed by biochemical analysis, revealed that uH2B directly activates methylation of H3 K79 by hDot1L. This effect is mediated through the catalytic domain of hDot1L, most likely through allosteric mechanisms. Furthermore, asymmetric incorporation of uH2B into dinucleosomes showed that the enhancement of methylation was limited to nucleosomes bearing uH2B. This work demonstrates a direct biochemical crosstalk between two modifications on separate histone proteins within a nucleosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007)
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000)
West, M. H. P. & Bonner, W. M. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 8, 4671–4680 (1980)
Robzyk, K., Recht, J. & Osley, M. A. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287, 501–504 (2000)
Kim, J., Hake, S. B. & Roeder, R. G. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell 20, 759–770 (2005)
Zhu, B. et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell 20, 601–611 (2005)
Ng, H. H., Xu, R. M., Zhang, Y. & Struhl, K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277, 34655–34657 (2002)
Briggs, S. D. et al. Trans-histone regulatory pathway in chromatin. Nature 418, 498 (2002)
Feng, Q. et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058 (2002)
Ng, H. H. et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16, 1518–1527 (2002)
van Leeuwen, F., Gafken, P. R. & Gottschling, D. E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756 (2002)
Henry, K. W. & Berger, S. L. Trans-tail histone modifications: wedge or bridge? Nature Struct. Biol. 9, 565–566 (2002)
Sun, Z. W. & Allis, C. D. Ubiquitylation of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 (2002)
Krogan, N. J. et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729 (2003)
Ezhkova, E. & Tansey, W. P. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of H3. Mol. Cell 13, 435–442 (2004)
Lee, J. S. et al. Histone crosstalk between H2B monoubiquitylation and H3 methylation mediated by COMPASS. Cell 131, 1084–1096 (2007)
Hwang, W. W. et al. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11, 251–266 (2003)
Wood, A. et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11, 267–274 (2003)
Muralidharan, V. & Muir, T. W. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nature Methods 3, 429–438 (2006)
Chatterjee, C., McGinty, R. K., Pellois, J. P. & Muir, T. W. Auxiliary-mediated site-specific peptide ubiquitylation. Angew. Chem. Int. Edn Engl. 46, 2814–2818 (2007)
Yan, L. Z. & Dawson, P. E. Synthesis of peptides and proteins without cysteine residues by native chemical ligation combined with desulfurization. J. Am. Chem. Soc. 123, 526–533 (2001)
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998)
Sawada, K. et al. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J. Biol. Chem. 279, 43296–43306 (2004)
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997)
Min, J., Feng, Q., Li, Z., Zhang, Y. & Xu, R. M. Structure of the catalytic domain of human Dot1L, a non-SET domain nucleosomal histone methyltransferase. Cell 112, 711–723 (2003)
Shahbazian, M. D., Zhang, K. & Grunstein, M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19, 271–277 (2005)
Garcia, B. A. et al. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 282, 7641–7655 (2007)
Zheng, C. & Hayes, J. J. Intra- and inter-nucleosomal protein–DNA interactions of the core histone tail domains in a model system. J. Biol. Chem. 26, 24217–24224 (2003)
Wang, S. S. et al. Facile synthesis of amino acid and peptide esters under mild conditions via cesium salts. J. Org. Chem. 42, 1286–1290 (1977)
Smith, A. B., Savinov, S. N., Manjappara, U. V. & Chaiken, I. M. Peptide-small molecule hybrids via orthogonal deprotection-chemoselective conjugation to cysteine-anchored scaffolds. Org. Lett. 4, 4041–4044 (2002)
Luger, K., Rechsteiner, T. J., Flaus, A. J., Waye, M. M. & Richmond, T. J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272, 301–311 (1997)
Pentelute, B. L. & Kent, S. B. Selective desulfurization of cysteine in the presence of Cys(Acm) in polypeptides obtained by native chemical ligation. Org. Lett. 9, 687–690 (2007)
Luger, K., Rechsteiner, T. J. & Richmond, T. J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119, 1–16 (1999)
Dorigo, B., Schalch, T., Bystricky, K. & Richmond, T. J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327, 85–96 (2003)
Owen-Hughes, T. et al. Analysis of nucleosome disruption by ATP-driven chromatin remodelling complexes. Methods Mol. Biol. 119, 319–331 (1999)
Ito, T. et al. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13, 1529–1539 (1999)
An, W., Kim, J. & Roeder, R. G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748 (2004)
Acknowledgements
We acknowledge H. Deng and J. Fernandez at The Rockefeller University Proteomics Resource Center for mass spectrometric analysis of methylated peptides. We thank T. J. Richmond for donating the 12_177_601 plasmid. We thank C. D. Allis for contributing the Xenopus histone plasmids for recombinant histone expression. We thank Y. Zhang for donating a plasmid containing hDot1L. We thank B. R. Rosenberg for assistance with phosphorimaging. We thank C. D. Allis, J. Tanny, and K. P. Chiang for discussions. This work was funded by the US National Institutes of Health. R.K.M. was supported by National Institutes of Health MSTP grant GM07739.
Author Contributions R.K.M., J.K. and C.C. did the experimental work; all authors performed project planning, data analysis and manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary information
The file contains Supplementary Figures S1-S11 with Legends. (PDF 974 kb)
Rights and permissions
About this article
Cite this article
McGinty, R., Kim, J., Chatterjee, C. et al. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453, 812–816 (2008). https://doi.org/10.1038/nature06906
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06906
This article is cited by
-
RNA modification: mechanisms and therapeutic targets
Molecular Biomedicine (2023)
-
H2B ubiquitination recruits FACT to maintain a stable altered nucleosome state for transcriptional activation
Nature Communications (2023)
-
Context-defined cancer co-dependency mapping identifies a functional interplay between PRC2 and MLL-MEN1 complex in lymphoma
Nature Communications (2023)
-
Proteasome inhibition targets the KMT2A transcriptional complex in acute lymphoblastic leukemia
Nature Communications (2023)
-
H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity
Nature Chemical Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.