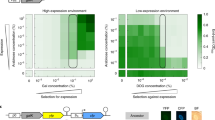
Abstract
Sequencing DNA from several organisms has revealed that duplication and drift of existing genes have primarily moulded the contents of a given genome. Though the effect of knocking out or overexpressing a particular gene has been studied in many organisms, no study has systematically explored the effect of adding new links in a biological network. To explore network evolvability, we constructed 598 recombinations of promoters (including regulatory regions) with different transcription or σ-factor genes in Escherichia coli, added over a wild-type genetic background. Here we show that ∼95% of new networks are tolerated by the bacteria, that very few alter growth, and that expression level correlates with factor position in the wild-type network hierarchy. Most importantly, we find that certain networks consistently survive over the wild type under various selection pressures. Therefore new links in the network are rarely a barrier for evolution and can even confer a fitness advantage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
ArrayExpress
Data deposits
Microarray data are MIAME-compliant and have been deposited at ArrayExpress http://www.ebi.ac.uk/microarray-as/aer/entry, accession E-MEXP-732.
References
Blattner, F. R. et al. The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1474 (1997)
Perez-Rueda, E. & Collado-Vides, J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 28, 1838–1847 (2000)
Salgado, H. et al. RegulonDB (version 3.2): Transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 29, 72–74 (2001)
Madan Babu, M. & Teichmann, S. A. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 31, 1234–1244 (2003)
Shen-Orr, S. S., Milo, R., Mangan, S. & Alon, U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature Genet. 31, 64–68 (2002)
Martinez-Antonio, A. & Collado-Vides, J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6, 482–489 (2003)
Salgado, H. et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 34, D394–D397 (2006)
Barabasi, A. L. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999)
Guelzim, N., Bottani, S., Bourgine, P. & Kepes, F. Topological and causal structure of the yeast transcriptional regulatory network. Nature Genet. 31, 60–63 (2002)
Albert, R., Jeong, H. & Barabasi, A. L. Error and attack tolerance of complex networks. Nature 406, 378–382 (2000)
Lynch, M. & Conery, J. S. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000)
Hooper, S. D. & Berg, O. G. On the nature of gene innovation: Duplication patterns in microbial genomes. Mol. Biol. Evol. 20, 945–954 (2003)
Teichmann, S. A., Park, J. & Chothia, C. Structural assignments to the Mycoplasma genitalium proteins show extensive gene duplications and domain rearrangements. Proc. Natl Acad. Sci. USA 95, 14658–14663 (1998)
Teichmann, S. A. & Babu, M. M. Gene regulatory network growth by duplication. Nature Genet. 36, 492–496 (2004)
Keseler, I. M. et al. EcoCyc: A comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33, D334–D337 (2005)
Mangan, M. W. et al. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 1831–1847 (2006)
Zaslaver, A. et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nature Methods 3, 623–628 (2006)
Pruss, B. M. & Matsumura, P. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J. Bacteriol. 178, 668–674 (1996)
Fong, S. S., Joyce, A. R. & Palsson, B. O. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 15, 1365–1372 (2005)
Dekel, E. & Alon, U. Optimality and evolutionary tuning of the expression level of a protein. Nature 436, 588–592 (2005)
Delaney, J. M., Ang, D. & Georgopoulos, C. Isolation and characterization of the Escherichia coli htrD gene, whose product is required for growth at high temperatures. J. Bacteriol. 174, 1240–1247 (1992)
Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6, 65–70 (1979)
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995)
Loewen, P. C., Hu, B., Strutinsky, J. & Sparling, R. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44, 707–717 (1998)
Cheville, A. M., Arnold, K. W., Buchrieser, C., Cheng, C. M. & Kaspar, C. W. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 62, 1822–1824 (1996)
Foster, J. W. & Moreno, M. Inducible acid tolerance mechanisms in enteric bacteria. Novartis Found. Symp. 221, 55–69; discussion 70–74 (1999)
Pratt, L. A., Hsing, W., Gibson, K. E. & Silhavy, T. J. From acids to osmZ: Multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20, 911–917 (1996)
Vidal, O. et al. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180, 2442–2449 (1998)
Prigent-Combaret, C. et al. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183, 7213–7223 (2001)
Cosma, C. L., Danese, P. N., Carlson, J. H., Silhavy, T. J. & Snyder, W. B. Mutational activation of the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope-associated stresses. Mol. Microbiol. 18, 491–505 (1995)
Romling, U., Bian, Z., Hammar, M., Sierralta, W. D. & Normark, S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180, 722–731 (1998)
Madan Babu, M., Teichmann, S. A. & Aravind, L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 358, 614–633 (2006)
Lozada-Chavez, I., Janga, S. C. & Collado-Vides, J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 34, 3434–3445 (2006)
Babu, M. M., Luscombe, N. M., Aravind, L., Gerstein, M. & Teichmann, S. A. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 14, 283–291 (2004)
Poelwijk, F. J., Kiviet, D. J. & Tans, S. J. Evolutionary potential of a duplicated repressor–operator pair: Simulating pathways using mutation data. PLoS Comput. Biol. 2, e58 (2006)
Hasty, J., McMillen, D. & Collins, J. J. Engineered gene circuits. Nature 420, 224–230 (2002)
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000)
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000)
Isalan, M., Lemerle, C. & Serrano, L. Engineering gene networks to emulate Drosophila embryonic pattern formation. PLoS Biol. 3, e64 (2005)
Isalan, M., Santori, M. I., Gonzalez, C. & Serrano, L. Localized transfection on arrays of magnetic beads coated with PCR products. Nature Methods 2, 113–118 (2005)
Sopko, R. et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21, 319–330 (2006)
Lange, R. & Hengge-Aronis, R. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol. 13, 733–743 (1994)
Solano, C. et al. Genetic analysis of Salmonella enteritidis biofilm formation: Critical role of cellulose. Mol. Microbiol. 43, 793–808 (2002)
Smyth, G. K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, article–3 (2004)
Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6, 65–70 (1979)
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995)
Acknowledgements
We thank A. Martinez Arias, J. Sharpe, M. Babu, P. Bork and B. Schoenwetter for critical reading of the manuscript; P. Ribeca for RegulonDB analysis; and B. Di Ventura and S. Martinez de Pablo for cloning assistance. C.H. and E.R. were funded by European Commission FP6 Netsensor Grant 012948.
Author Contributions M.I., C.L., K.M., P.B., C.H. and M.G.-C. carried out experiments. E.R. and C.L. did computational analysis. M.I., C.L. and L.S. conceived experiments. M.I. and L.S. supervised experiments.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-9 with Legends, Supplementary Methods and Supplementary Notes. (PDF 2727 kb)
Supplementary Data 1
The file contains Supplementary Data 1 with data for growth and GFP under 5 conditions. (XLS 242 kb)
Supplementary Data 2
The file contains Supplementary Data 2 with data for RTqPCR and raw growth signatures. (XLS 12012 kb)
Supplementary Data 3
The file contains Supplementary Data 3 with data for DNA chip analysis. (XLS 18086 kb)
Rights and permissions
About this article
Cite this article
Isalan, M., Lemerle, C., Michalodimitrakis, K. et al. Evolvability and hierarchy in rewired bacterial gene networks. Nature 452, 840–845 (2008). https://doi.org/10.1038/nature06847
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06847
This article is cited by
-
Robustness and innovation in synthetic genotype networks
Nature Communications (2023)
-
Genome-wide promoter responses to CRISPR perturbations of regulators reveal regulatory networks in Escherichia coli
Nature Communications (2023)
-
A quantitative method for proteome reallocation using minimal regulatory interventions
Nature Chemical Biology (2020)
-
Evolutionary potential of transcription factors for gene regulatory rewiring
Nature Ecology & Evolution (2018)
-
Network analyses in systems biology: new strategies for dealing with biological complexity
Synthese (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.