Abstract
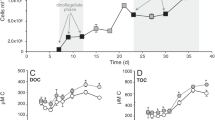
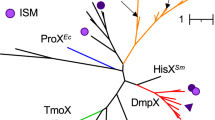
Sulphur is a universally required cell nutrient found in two amino acids and other small organic molecules. All aerobic marine bacteria are known to use assimilatory sulphate reduction to supply sulphur for biosynthesis, although many can assimilate sulphur from organic compounds that contain reduced sulphur atoms1,2,3. An analysis of three complete ‘Candidatus Pelagibacter ubique’ genomes, and public ocean metagenomic data sets, suggested that members of the ubiquitous and abundant SAR11 alphaproteobacterial clade are deficient in assimilatory sulphate reduction genes. Here we show that SAR11 requires exogenous sources of reduced sulphur, such as methionine or 3-dimethylsulphoniopropionate (DMSP) for growth. Titrations of the algal osmolyte DMSP in seawater medium containing all other macronutrients in excess showed that 1.5 × 108 SAR11 cells are produced per nanomole of DMSP. Although it has been shown that other marine alphaproteobacteria use sulphur from DMSP in preference to sulphate1,2, our results indicate that ‘Cand. P. ubique’ relies exclusively on reduced sulphur compounds that originate from other plankton.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kiene, R. P., Linn, L. J., Gonzalez, J., Moran, M. A. & Bruton, J. A. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65, 4549–4558 (1999)
Cuhel, R. L., Taylor, C. D. & Jannasch, H. W. Assimilatory sulfur metabolism in marine microorganisms: sulfur metabolism, protein synthesis, and growth of Alteromonas luteo-violaceus and Pseudomonas halodurans during perturbed batch growth. Appl. Environ. Microbiol. 43, 151–159 (1982)
Cottrell, M. T. & Kirchman, D. L. Natural assemblages of marine proteobacteria and members of the Cytophaga–Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66, 1692–1697 (2000)
Morris, R. M. et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810 (2002)
Street, J. H. & Paytan, A. Iron, phytoplankton growth, and the carbon cycle. Met. Ions Biol. Syst. 43, 153–193 (2005)
Schut, F. et al. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59, 2150–2160 (1993)
Neumann, S., Wynen, A., Truper, H. G. & Dahl, C. Characterization of the cys gene locus from Allochromatium vinosum indicates an unusual sulfate assimilation pathway. Mol. Biol. Rep. 27, 27–33 (2000)
Ruckert, C. et al. Functional genomics and expression analysis of the Corynebacterium glutamicum fpr2-cysIXHDNYZ gene cluster involved in assimilatory sulphate reduction. BMC Genomics 6, 121 (2005)
Alm, E. J. et al. The MicrobesOnline Web site for comparative genomics. Genome Res. 15, 1015–1022 (2005)
Berndt, C. et al. Characterization and reconstitution of a 4Fe-4S adenylyl sulfate/phosphoadenylyl sulfate reductase from Bacillus subtilis. J. Biol. Chem. 279, 7850–7855 (2004)
Heinzinger, N. K., Fujimoto, S. Y., Clark, M. A., Moreno, M. S. & Barrett, E. L. Sequence analysis of the phs operon in Salmonella typhimurium and the contribution of thiosulfate reduction to anaerobic energy metabolism. J. Bacteriol. 177, 2813–2820 (1995)
Malmstrom, R. R., Kiene, R. P., Cottrell, M. T. & Kirchman, D. L. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl. Environ. Microbiol. 70, 4129–4135 (2004)
Howard, E. C. et al. Bacterial taxa that limit sulfur flux from the ocean. Science 314, 649–652 (2006)
Todd, J. D. et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315, 666–669 (2007)
Kiene, R. P. & Linn, L. J. The fate of dissolved dimethylsulfoniopropionate (DMSP) in seawater: Tracer studies using S-35-DMSP. Geochim. Cosmochim. Acta 64, 2797–2810 (2000)
Nicastro, D. et al. Three-dimensional structure of the tiny bacterium Pelagibacter ubique studied by cryo-electron tomography. Microsc. Microanal. 12, 180–181 (2006)
Simo, R., Archer, S. D., Pedros-Alio, C., Gilpin, L. & Stelfox-Widdicombe, C. E. Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol. Oceanogr. 47, 53–61 (2002)
Zubkov, M. V. et al. Rapid turnover of dissolved DMS and DMSP by defined bacterioplankton communities in the stratified euphotic zone of the North Sea. Deep-Sea Res. II 49, 3017–3038 (2002)
Lippert, K. D. & Pfennig, N. Utilisation of molecular hydrogen by Chlorobium thiosulfatophilum. Growth and CO2-fixation. Arch. Mikrobiol. 65, 29–47 (1969)
Glaeser, J. & Overmann, J. Selective enrichment and characterization of Roseospirillum parvum, gen. nov. and sp. nov., a new purple nonsulfur bacterium with unusual light absorption properties. Arch. Microbiol. 171, 405–416 (1999)
Ventura, S., De Philippis, R., Materassi, R. & Balloni, W. Two halophilic Ectothiorhodospira strains with unusual morphological, physiological and biochemical characters. Arch. Microbiol. 149, 273–279 (1988)
Daniels, L., Belay, N. & Rajagopal, B. S. Assimilatory reduction of sulfate and sulfite by methanogenic bacteria. Appl. Environ. Microbiol. 51, 703–709 (1986)
van Ham, R. C. et al. Reductive genome evolution in Buchnera aphidicola. Proc. Natl Acad. Sci. USA 100, 581–586 (2003)
Hou, S. et al. Genome sequence of the deep-sea gamma-proteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy. Proc. Natl Acad. Sci. USA 101, 18036–18041 (2004)
Giovannoni, S. J. et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245 (2005)
Rappe, M. S., Connon, S. A., Vergin, K. L. & Giovannoni, S. J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633 (2002)
Connon, S. A. & Giovannoni, S. J. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68, 3878–3885 (2002)
Stingl, U., Tripp, H. J. & Giovannoni, S. J. Improvements of high-throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site. ISME J 1, 361–371 (2007)
Peterson, J. D., Umayam, L. A., Dickinson, T., Hickey, E. K. & White, O. The comprehensive microbial resource. Nucleic Acids Res. 29, 123–125 (2001)
Bernal, A., Ear, U. & Kyrpides, N. Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 29, 126–127 (2001)
Acknowledgements
We thank U. Stingl, C. Carlson and A. Treusch for discussions, R. Kiene for 35S[DMSP], and K. Vergin for assistance with radiotracers. This work was supported by grants from the Marine Microbiology Initiative of the Gordon and Betty Moore Foundation and the National Science Foundation.
Author Contributions H.J.T. conducted genome analysis, adaptation of Guava EasyCyte to marine picoplankton counting, initial experimental design, preliminary data collection and analysis, and wrote the manuscript. J.B.K. set up, performed, and collected data for many culture experiments, and assisted in EasyCyte adaptations. J.W.H.D. provided DMSP and DMSP measurements. L.J.W. performed metagenomics. S.J.G. initiated the study, suggested candidate nutrients for testing, and with M.S.S. suggested final experiments and data presentation. All authors reviewed the manuscript before submission.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
The file contains Supplementary Methods, Supplementary Notes, Supplementary Figures 1-4 with Legends and Supplementary Table 1 with Legend. The Supplementary Methods describe the search for sulphate reduction genes. The Supplementary Notes describe cell size and morphology, identify putative genes and metabolic pathways for sulphur metabolism, and demonstrate the infeasibility of 35SO4 uptake experiments, other sources of reduced sulfur in the oceanic water column, and consistency of cell morphology and size. The Supplementary Figures and Tables are included within the notes. (PDF 195 kb)
Rights and permissions
About this article
Cite this article
Tripp, H., Kitner, J., Schwalbach, M. et al. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452, 741–744 (2008). https://doi.org/10.1038/nature06776
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06776
This article is cited by
-
Salinity-controlled distribution of prokaryotic communities in the Arctic sea-ice melt ponds
World Journal of Microbiology and Biotechnology (2024)
-
Taxonomic and environmental distribution of bacterial amino acid auxotrophies
Nature Communications (2023)
-
Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria
The ISME Journal (2023)
-
Viral infection switches the balance between bacterial and eukaryotic recyclers of organic matter during coccolithophore blooms
Nature Communications (2023)
-
Ecophysiology and genomics of the brackish water adapted SAR11 subclade IIIa
The ISME Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.