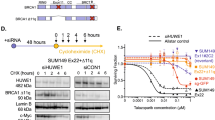
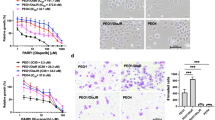
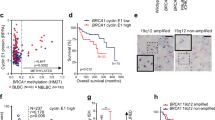
Abstract
Ovarian carcinomas with mutations in the tumour suppressor BRCA2 are particularly sensitive to platinum compounds1. However, such carcinomas ultimately develop cisplatin resistance. The mechanism of that resistance is largely unknown2. Here we show that acquired resistance to cisplatin can be mediated by secondary intragenic mutations in BRCA2 that restore the wild-type BRCA2 reading frame. First, in a cisplatin-resistant BRCA2-mutated breast-cancer cell line, HCC1428, a secondary genetic change in BRCA2 rescued BRCA2 function. Second, cisplatin selection of a BRCA2-mutated pancreatic cancer cell line, Capan-1 (refs 3, 4), led to five different secondary mutations that restored the wild-type BRCA2 reading frame. All clones with secondary mutations were resistant both to cisplatin and to a poly(ADP-ribose) polymerase (PARP) inhibitor (AG14361). Finally, we evaluated recurrent cancers from patients whose primary BRCA2-mutated ovarian carcinomas were treated with cisplatin. The recurrent tumour that acquired cisplatin resistance had undergone reversion of its BRCA2 mutation. Our results suggest that secondary mutations that restore the wild-type BRCA2 reading frame may be a major clinical mediator of acquired resistance to platinum-based chemotherapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Yuan, S. S. et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59, 3547–3551 (1999)
Agarwal, R. & Kaye, S. B. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nature Rev. Cancer 3, 502–516 (2003)
Goggins, M. et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 56, 5360–5364 (1996)
Abbott, D. W., Freeman, M. L. & Holt, J. T. Double-strand break repair deficiency and radiation sensitivity in BRCA2 mutant cancer cells. J. Natl Cancer Inst. 90, 978–985 (1998)
Li, A. J. & Karlan, B. Y. Genetic factors in ovarian carcinoma. Curr. Oncol. Rep. 3, 27–32 (2001)
van Asperen, C. J. et al. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J. Med. Genet. 42, 711–719 (2005)
Howlett, N. G. et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297, 606–609 (2002)
Moynahan, M. E., Pierce, A. J. & Jasin, M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7, 263–272 (2001)
Neuhausen, S. L. & Marshall, C. J. Loss of heterozygosity in familial tumors from three BRCA1-linked kindreds. Cancer Res. 54, 6069–6072 (1994)
Collins, N. et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12–13. Oncogene 10, 1673–1675 (1995)
Gudmundsson, J. et al. Different tumor types from BRCA2 carriers show wild-type chromosome deletions on 13q12–q13. Cancer Res. 55, 4830–4832 (1995)
Bhattacharyya, A., Ear, U. S., Koller, B. H., Weichselbaum, R. R. & Bishop, D. K. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275, 23899–23903 (2000)
Tutt, A. N. et al. Exploiting the DNA repair defect in BRCA mutant cells in the design of new therapeutic strategies for cancer. Cold Spring Harb. Symp. Quant. Biol. 70, 139–148 (2005)
Foulkes, W. D. BRCA1 and BRCA2: chemosensitivity, treatment outcomes and prognosis. Fam. Cancer 5, 135–142 (2006)
Boyd, J. et al. Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. J. Am. Med. Assoc. 283, 2260–2265 (2000)
Hirschhorn, R. In vivo reversion to normal of inherited mutations in humans. J. Med. Genet. 40, 721–728 (2003)
Hamanoue, S. et al. Myeloid lineage-selective growth of revertant cells in Fanconi anaemia. Br. J. Haematol. 132, 630–635 (2006)
Xia, B. et al. Fanconi anaemia is associated with a defect in the BRCA2 partner PALB2. Nature Genet. 39, 159–161 (2007)
Gazdar, A. F. et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int. J. Cancer 78, 766–774 (1998)
Neuhausen, S. et al. Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nature Genet. 13, 126–128 (1996)
Saeki, H. et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc. Natl Acad. Sci. USA 103, 8768–8773 (2006)
Wu, K. et al. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 65, 417–426 (2005)
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005)
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005)
Jacquemont, C. & Taniguchi, T. Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res. 67, 7395–7405 (2007)
Tutt, A. et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 20, 4704–4716 (2001)
Hilton, J. L. et al. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J. Natl. Cancer Inst. 94, 1396–1406 (2002)
Ikeda, H. et al. Genetic reversion in an acute myelogenous leukemia cell line from a Fanconi anemia patient with biallelic mutations in BRCA2. Cancer Res. 63, 2688–2694 (2003)
Wiegant, W. W., Overmeer, R. M., Godthelp, B. C., van Buul, P. P. & Zdzienicka, M. Z. Chinese hamster cell mutant, V-C8, a model for analysis of Brca2 function. Mutat. Res. 600, 79–88 (2006)
Gorre, M. E. et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293, 876–880 (2001)
Taniguchi, T. et al. Disruption of the Fanconi anemia–BRCA pathway in cisplatin-sensitive ovarian tumors. Nature Med. 9, 568–574 (2003)
Garcia-Higuera, I. et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7, 249–262 (2001)
Taniguchi, T. et al. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109, 459–472 (2002)
Chirnomas, D. et al. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol. Cancer Ther. 5, 952–961 (2006)
Naf, D., Kupfer, G. M., Suliman, A., Lambert, K. & D’Andrea, A. D. Functional activity of the fanconi anemia protein FAA requires FAC binding and nuclear localization. Mol. Cell. Biol. 18, 5952–5960 (1998)
Skalitzky, D. J. et al. Tricyclic benzimidazoles as potent poly(ADP-ribose) polymerase-1 inhibitors. J. Med. Chem. 46, 210–213 (2003)
Yamashita, T., Barber, D. L., Zhu, Y., Wu, N. & D’Andrea, A. D. The Fanconi anemia polypeptide FACC is localized to the cytoplasm. Proc. Natl Acad. Sci. USA 91, 6712–6716 (1994)
Bruun, D. et al. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair 2, 1007–1013 (2003)
Trask, B. J. in Genome Analysis: A Laboratory Manual (eds Birren, B. et al.), vol. 4 303–413 (Cold Spring Harbor Laboratory Press, New York, 1999)
Acknowledgements
We thank M.C. King and C. W. Drescher for discussions, B. Trask for overseeing the FISH analyses in her laboratory and for comments on the manuscript, J.W. Huang for comments on the manuscript, and M. Hoatlin and K. Polyak for reagents. We thank Pfizer for AG14361. We thank the Pacific Ovarian Cancer Research Consortium (supported by a Specialized Program of Research Excellence in Ovarian Cancer) for clinical specimens. This work was supported by grants from the National Institutes of Health/National Cancer Institute (to T.T. and E.M.S.), the Searle Scholars Program, the V Foundation and the Hartwell Innovation Fund (to T.T.), the L&S Milken Foundation (to B.Y.K.), the American Cancer Society California Division-Early Detection Professorship (to B.Y.K.), start-up funds from the Fred Hutchinson Cancer Research Center (to T.T.) and a gift from the Yvonne Betson Trust (to E.M.S.).
Author Contributions W.S. performed most of the experiments. E.M.S., B.Y.K. and N.U. provided clinical samples and expertise on ovarian cancer. E.M.S. performed laser capture microdissection and DNA extractions. M.K.A., D.J.F. and F.J.C. performed homologous recombination assays. J.H. sequenced BRCA2 in HCC1428. C.F. performed FISH analysis. E.V. performed the siRNA experiments shown in Fig. 1f, g. C.J. performed the PARP inhibitor sensitivity assays in Fig. 3c. T.T., W.S. and E.M.S. wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
The file contains Supplementary Figures S1-S9 with Legends and Supplementary Tables S1-S5. (PDF 4082 kb)
Rights and permissions
About this article
Cite this article
Sakai, W., Swisher, E., Karlan, B. et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116–1120 (2008). https://doi.org/10.1038/nature06633
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06633
This article is cited by
-
Convergent evolution of BRCA2 reversion mutations under therapeutic pressure by PARP inhibition and platinum chemotherapy
npj Precision Oncology (2024)
-
Metastatic lung adenocarcinoma with BRCA2 mutation and longstanding disease control on olaparib, developing triple negative breast adenocarcinoma with additional BRCA2 reversion mutation: a case report
Journal of Medical Case Reports (2023)
-
Dramatic, durable response to therapy in gBRCA2-mutated pancreas neuroendocrine carcinoma: opportunity and challenge
npj Precision Oncology (2023)
-
PARP inhibitors: enhancing efficacy through rational combinations
British Journal of Cancer (2023)
-
Single-cell characterization of step-wise acquisition of carboplatin resistance in ovarian cancer
npj Systems Biology and Applications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.