Abstract
A principal question in MAP kinase (MAPK/MPK) cascade signalling is how similar components dictate different specificity in the information-processing machineries from yeast to humans and plants. In Arabidopsis, how MPK3/6 modulates distinct outputs in diverse signal transduction pathways remains elusive. By combining systematic cellular and genetic screens, here we uncover a previously unexpected MKK9–MPK3/MPK6 cascade promoting ethylene-insensitive 3 (EIN3)-mediated transcription in ethylene signalling. The mkk9 mutant exhibits a broad spectrum of moderate ethylene-insensitive phenotypes, and translocated MKK9 governs nuclear signalling downstream of receptors. Breaking a linear model and conventional MAPK signalling, ethylene inactivates the negative regulator constitutive triple response 1 (CTR1, a Raf-like MAPK kinase kinase (MAPKKK)) to activate the positive MKK9–MPK3/6 cascade. The bifurcate and antagonistic CTR1 and MKK9 pathways are both critical in determining ethylene-signalling specificity through two MAPK phosphorylation sites with opposite effects on EIN3 stability. The results suggest a new paradigm for linking intertwined MAPK cascades to control quantitative responses and specificity in signalling networks.
Similar content being viewed by others
Main
Ethylene (C2H4) was the first example of a gaseous signalling molecule in biological systems, discovered more than a century ago1. As a major plant hormone, it controls essential physiological processes, including: germination; root, shoot and flower development; stress, defence and glucose responses; fruit ripening; and senescence1,2,3,4,5. Extensive genetic analysis of ethylene signal transduction in Arabidopsis has established a linear pathway connecting five receptors2,3,4,5,6,7 to a single negative regulator, CTR1 (ref. 8), and two key downstream positive components, EIN2 (ref. 9) and EIN3 (ref. 10). CTR1 encodes a putative Raf-like MAPKKK and interacts with the ethylene response 1 (ETR1) receptor, but its biochemical activity and molecular actions are unclear8,11,12. EIN3 and its closely related EIN3-LIKE1 (EIL1) are plant-specific nuclear transcription factors that initiate downstream transcriptional cascades for ethylene responses10,13,14,15. The identification of ethylene receptor, CTR1, EIN2 and EIN3 orthologues in diverse plant species3 suggests the evolutionary conservation of ethylene signalling. However, it remains unknown how CTR1 functions as a MAPKKK to regulate downstream positive signalling components in the nucleus.
EIN3 interacts with two F-box proteins (EBF1 and EBF2) and is degraded by the 26S proteasome5,16,17,18,19. Ubiquitin/proteasome-dependent protein degradation mediated by specific F-box proteins of the conserved SCF (SKP1/cullin/F-box protein) E3 ubiquitin ligase complexes has emerged as a universal mechanism in response to multiple plant hormones, including auxin, gibberellin, abscisic acid, jasmonate and ethylene20,21. Understanding the distinct upstream signalling pathways that quantitatively control EIN3 and other transcription regulators through F-box protein-mediated degradation is a major interest in biology.
MAPK cascades are pivotal signalling modules controlling diverse signal transduction pathways in eukaryotes. A main question in MAPK cascade signalling has been how similar components control different biological responses22,23,24,25,26,27. In yeast and mammals, the specificity for distinct signalling pathways with shared components is largely determined by scaffolding proteins and specific MAPKs25,26,27. In plants, more complex roles of MAPKs have emerged. For instance, diverse hormones, stresses, microbial elicitors and developmental processes all activate the same conserved MPK3 and MPK6 in Arabidopsis and their orthologues in other plant species22,23,24,28,29. In ethylene responses, genetic and biochemical data present contradicting negative and positive roles of MAPK signalling8,30,31,32. It is unclear whether and how MAPK cascades are involved in the current linear genetic framework of the ethylene signal transduction pathway3,4,5. Adding to the complexity, Arabidopsis MPK6 is also shown to phosphorylate and stabilize ethylene biosynthetic enzymes, 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS2/6), to promote ethylene synthesis33. Plant genomes encode the most extended families of putative MAPK cascade genes in eukaryotes22,24. Combining cellular and genetic screens with genomic information, we have initiated systematic and quantitative analyses of Arabidopsis MAPK cascades. Here we report the unexpected discovery of an MKK9–MPK3/6 cascade that positively modulates nuclear EIN3 stability. Remarkably, CTR1 acts as an unconventional MAPKKK, blocking MKK9–MPK3/MPK6 activation and simultaneously enhancing EIN3 degradation by distinct MPK phosphorylation. Our results illuminate new molecular mechanisms for the control of transcription-factor stability by intertwined MAPK cascades to achieve quantitative signalling specificity in eukaryotes.
Screens for MPKs and MKKs in ethylene signalling
To assess critically the role and molecular identity of MAPK cascades in ethylene signalling, we have initiated an integrative approach by combining molecular, genomic, biochemical and genetic tools. To increase the sensitivity of the MAPK screen, we used ctr1-1 (ctr1) protoplasts with no endogenous CTR1 activity in the background15,30,31,34,35,36. Unexpectedly, constitutively active CTR1 (CTR1a) clearly repressed MPK1, MPK3 and MPK6 activities without altering protein expression based on an immunocomplex MAPK assay34,35,36 (Fig. 1a). The result raised a possibility that the putative CTR1 MAPKKK could negatively regulate downstream MAPK activities, which could play a positive role in ethylene signalling22,30,31. Because little MPK1 but abundant MPK3 and MPK6 transcripts were detected in protoplasts and leaves (Supplementary Fig. 1a), MPK3 and MPK6 are probably the physiological MAPKs in ethylene signalling.
a, Specific MAPKs are activated in the ctr1 protoplasts but inactivated by CTR1a. MAPK activity and protein expression (MPK–HA or CTR1a–Flag) are shown. The experiments were repeated twice. b, Constitutively active MKK4a, MKK5a, MKK7a and MKK9a activate MPK3 and MPK6 in WT protoplasts. c, Transient expression of WT MKK7 or MKK9 preferentially increases MPK3 and MPK6 activity (act.) in ctr1 protoplasts. Empty vector (-) was used as a DNA transfection control. Protein expression (MKK–MYC or MPK–HA) is shown.
We next attempted to identify the MAPKKs (MKKs) that could activate MPK3/6 in ethylene signalling. There are ten putative MKK genes in the Arabidopsis genome22,24, and all except MKK8 and MKK10 are expressed in mesophyll cells. We performed a primary screen to identify constitutively active MKK (MKKa) that could activate co-expressed MPK3/6 in wild-type (WT) protoplasts. The MKKa constructs were generated by site-directed mutagenesis to convert threonine/serine to aspartic acid/glutamic acid mimicking phosphorylation36 and activation by upstream MAPKKKs. In addition to MKK4a and MKK5a36, MKK7a and MKK9a activated MPK3/6 based on an immunocomplex MAPK assay (Fig. 1b). Interestingly, overexpressing WT MKK7 and MKK9 specifically and preferentially activated the epitope-tagged MPK3/6 (Fig. 1c) or endogenous MAPKs corresponding to the size of MPK3/6 (Supplementary Fig. 1b) in ctr1 protoplasts lacking the key negative regulator CTR1. Wild-type MKK4 and MKK5 did not activate MPK3/6 in WT or ctr1 protoplasts (Fig. 1c and Supplementary Fig. 1c). Although MKK4/5 and MKK7/9 could all activate MPK3/6, they appeared to have differential functions in MAPK signalling networks. The CTR1 MAPKKK could act unconventionally as a direct or indirect negative regulator of the MKK7/9–MPK3/6 cascade.
MKK9 activates EIN3-mediated transcription in ctr1
To connect this novel and positive MAPK cascade to the key and specific nuclear transcription factor EIN3 in ethylene signalling, we conducted a second MKK screen using an early ethylene-responsive luciferase (LUC) reporter directly targeted by EIN3 for transcription activation15. The reporter carrying four synthetic copies of the defined EIN3 binding site (EBS)10,13,15 was highly and specifically activated by EIN3 co-expression within 3 h in the transient assay (Fig. 2a). The short timeframe ensured the activation of only the earliest and direct EIN3 target genes and avoided secondary responses. The promoter of an early ethylene response gene ERF5, but not other promoters inducible by flg22 (WRKY29)36, abscisic acid (RD29A)35 and auxin (GH3)34,35, was also activated by co-expressed EIN3, confirming the specificity of the assay (Fig. 2a). Furthermore, EBS-LUC responded to ethylene in WT but not in ethylene-insensitive ein2 protoplasts lacking EIN3 (ref. 17) (Fig. 2b). Correlated with the CTR1a repression of MPK3/6 activities (Fig. 1a), CTR1a but not inactive InCTR1K579M reduced EBS-LUC activity in ctr1 protoplasts (Supplementary Fig. 1d).
a, EIN3 specifically induces EBS-LUC and ERF5-LUC activities in WT protoplasts. b, C2H4 (10 p.p.m.) induces EBS-LUC activity in WT but not in ein2-1 protoplasts. c, WT MKK7 or MKK9 specifically enhances EBS-LUC activities in ctr1 protoplasts. d, Constitutively active MKK7a or MKK9a further enhances EBS-LUC activities in ctr1 protoplasts. Protein expression (MKK–MYC) is shown. Error bars, s.d. (n = 3). All experiments were repeated three times with similar results.
We performed a systematic screen based on quantitative EBS-LUC response to identify WT MKKs that could enhance ethylene signalling in ctr1 protoplasts. In agreement with the specific MPK3/6 activation in ctr1 protoplasts (Fig. 1c), expression of WT MKK7 or MKK9 uniquely enhanced EBS-LUC activity (Fig. 2c). The constitutively active MKK7a or MKK9a displayed even greater ability to activate EBS-LUC (Fig. 2d). Arabidopsis MKK4a/5a and tobacco NtMEKDD activate MPK6, which phosphorylates and enhances cytosolic ACS2/6 stability to elevate ethylene levels33. The differential MKK4a/5a–MPK3/6 and MKK7a/9a–MPK3/6 effect on the early ethylene reporter gene activation in ctr1 suggests signalling specificity downstream of ethylene perception, probably in the nucleus. Because the ctr1 mutant phenotype is not as severe as etr1 ers1 or ebf1 ebf2 double mutants16,18,19,37,38 and can further respond to ethylene39, this positive MAPK cascade may represent an additional regulatory pathway between the receptors and the F-box proteins. Although MKK7 and MKK9 are highly homologous and showed similar activities in the MAPK and reporter activation, the steady-state MKK9 transcript level was much higher in protoplasts and leaves (Supplementary Fig. 1e). Thus, MKK9 is likely to play a predominant role in ethylene signalling in mesophyll cells. Consistently, the activated MKK9 but not the MKK9 kinase mutant immunoprecipitated from protoplasts could directly phosphorylate in vitro GST–MPK3/6 produced from Escherichia coli without autophosphorylation36 (Supplementary Fig. 2).
mkk9 exhibits diverse ethylene-insensitive phenotypes
To obtain genetic evidence to independently evaluate the surprising and specific positive role of MKK9 in ethylene signalling, we performed quantitative ethylene response screens of mkk loss-of-function mutants collected from various transferred DNA (T-DNA) insertion resources40,41 (Supplementary Figs 3–5 and Supplementary Table 1). The WT and various mutant plants were grown under the same condition and harvested at the same time to minimize plant-to-plant variations. As shown in Fig. 3a, only the etiolated mkk9 seedlings displayed ethylene insensitivity in hypocotyl similar to ein3 in the presence of 0.5–1 μM of ACC3,4,5,10,14,19. The ethylene-insensitive phenotype was similar in two distinct mkk9 alleles from different sources (Supplementary Fig. 5). We further examined the specific role of MKK9 in ethylene signalling by comparing the phenotypes of mkk9-1 and ein3 in five additional assays10,15,42,43. Both mkk9 and ein3 displayed similar hypersensitivity to glucose and salt (Fig. 3b, c). The link between ethylene and glucose or salt sensitivity was further supported by the hypersensitive phenotypes of other stronger ethylene-insensitive mutants, such as etr1 and ein2 (refs 42, 43). Both mkk9 and ein3 leaves showed little chlorophyll degradation induced by ACC in the dark10 (Supplementary Fig. 6a), and were relatively resistant to ethylene inhibition of leaf and petiole elongation under light10 (Supplementary Fig. 6b). Importantly, ethylene activation of immediate early marker genes, ERF1 and ERF5, was abolished in mkk9 and ein3 leaves (Fig. 3d). These marker genes are direct EIN3 targets13,15 (Fig. 2a) and are specific indicators of early ethylene signalling. The inability of mkk9 to respond to exogenous ethylene in early transcription activation ruled out the possibility that the mkk9 phenotypes were due to lower endogenous ethylene biosynthesis. The results also provided important evidence to uncouple the MAPK regulation of ethylene signalling from ethylene synthesis.
a, The mkk9 and ein3 seedlings exhibit ethylene insensitivity in hypocotyl elongation at 0.5–1 µM ACC. Error bars, s.d. (n = 20). Asterisks indicate differences between WT and mutant with statistical significance at *P < 0.05, **P < 0.01 and ***P < 0.001 (t-test). b, c, The seedlings of mkk9 and other ethylene-insensitive mutants show glucose (b) and salt (c) hypersensitivity. d, Immediate early gene induction by C2H4 (5 p.p.m.) is defective in mkk9 and ein3 leaves measured by qRT–PCR. Error bars, s.d. (n = 3). e, Endogenous MAPKs activated by ACC in WT, mpk3 and mpk6 leaves. Leaves were treated with 200 μM ACC for 1 h34,35,36. Rubisco (anti-RBC), loading control.
To assess whether the ethylene insensitivity of mkk9 is due to the loss of the MKK9-dependent MPK3/6 activation in ethylene signalling, ACC activation of MAPK activity was measured in detached leaves after assay optimization (Supplementary Fig. 7a). Two endogenous MAPKs were activated in WT but not in mkk9 after ACC feeding through the petiole to minimize mechanical stress during the treatment (Fig. 3e and Supplementary Fig. 7a). Maximal MAPK activities were detected at 1 h because of the time required for ACC uptake and conversion to ethylene in vivo. We used the loss-of-function mpk3 and mpk6 single mutants33 to verify that the ACC- and MKK9-dependent activation of MAPKs were indeed MPK3 and MPK6 (Fig. 3e). Our results are consistent with previous independent studies showing that ethylene or ACC activated MAPK30,31. Interestingly, MPK6 activation by ACC was elevated in mpk3, whereas the activation of MPK3 was greatly enhanced in mpk6. The results suggest that MPK3 and MPK6 play redundant roles and can compensate for the loss of each other in ethylene signalling (Fig. 3e). Using the virus-induced gene silence (VIGS) method44 to bypass embryo lethality29, we showed that the inductions of ERF1 and ERF5 were significantly diminished in the mpk3 mpk6 double mutants (Supplementary Fig. 7b). The combined genetic analyses of mkk9 and mpk3 mpk6 mutant plants provide compelling in vivo evidence to support their specific roles in ethylene signalling.
MKK9a promotes constitutive ethylene signalling
To substantiate the role of MKK9 in ethylene signalling, we generated transgenic Arabidopsis plants expressing the constitutively active MKK9a in WT or different ethylene-insensitive mutants. Transgenic lines were first screened for equal MKK9a transgene expression in the WT, etr1 and ein2 background (Supplementary Fig. 8a) without overt cell death before the phenotypic analyses. MKK9a transgenic plants showed constitutive ethylene phenotypes in the WT background (Fig. 4a, b). Similar ethylene phenotypes have been observed in multiple receptor mutants7,37,38 or the double mutant ebf1 ebf2 accumulating EIN3 (refs 16, 18, 19). Moreover, ERF1 and ERF5 were highly activated by MKK9a in the transgenic Arabidopsis (Fig. 4c).
a, MKK9a confers constitutive ethylene responses. Ag+ (100 µM). b, Adult 28-day-old MKK9a transgenic plants exhibit a constitutive ethylene phenotype. Scale bar, 10 mm. c, MKK9a promotes immediate early ethylene gene expression measured by qRT–PCR. Error bars, s.d. (n = 3). d, MKK9a action is blocked in ein2 but not in etr1 seedlings. Control (Cont) etr1 or ein2 seedlings do not carry MKK9a. e, The mkk9 etiolated seedlings are complemented by MKK9 (MKK9/mkk9). f, Induction of ethylene response genes measured by qRT–PCR is restored in the MKK9-complemented lines (MKK9/mkk9). Error bars, s.d. (n = 3).
To place MKK9a in the genetically established ethylene-signalling pathway2,3,4,5, we analysed the effects of MKK9a in the ethylene-insensitive etr1 and ein2 mutants. The MKK9a phenotypes, including etiolated seedling responses and gene activation, were not blocked by etr1 with little ethylene perception (Fig. 4c, d). The results were consistent with the insensitivity of MKK9a seedling phenotypes to an ethylene receptor antagonist Ag+ (ref. 17) (Fig. 4a), suggesting that MKK9a acted downstream of the ethylene receptors but not simply enhancing ethylene synthesis. In contrast, the MKK9a phenotypes were diminished in the ein2 mutant (Fig. 4c, d). A straightforward genetic model would place MKK9 upstream of EIN2, analogous to the common interpretation of the ctr1 ein2 double mutant2,3,4,5. However, it is equally possible that the reduced effects of MKK9a could be explained by the absence of EIN3 as the MKK9 cascade target in the ein2 mutant17, in which EIN3 overexpression could overcome its defect10. The gain-of-function analyses of MKK9a in transgenic plants provided additional in vivo evidence to support its role in ethylene signalling. The mkk9 mutant was complemented (Fig. 4e, f) with WT MKK9 (Supplementary Fig. 8b). Although the etiolated ctr1 and the ctr1 mkk9 double mutant seedlings looked similar, the elongated light-grown ctr1 seedling phenotype9 was diminished in the ctr1 mkk9 double mutant in the absence or presence of ACC (Supplementary Fig. 9), supporting the idea that MKK9 acted downstream of CTR1.
Dual phosphorylation modulates EIN3 stability
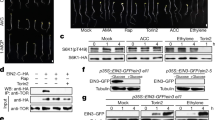
We next examined how MKK9–MPK3/6 cascades might regulate EIN3 in ethylene signalling15,16,17,18. Interestingly, an MKK-specific inhibitor, U0126, blocked EIN3 accumulation induced by ACC (Fig. 5a), and phosphatase caused EIN3 band shift, suggesting EIN3 phosphorylation in seedlings (Fig. 5b). Constantly, the constitutively active MKK9a–GFP but not MKK4a–GFP preferentially localized in the nucleus (Fig. 5c), whereas MPK6 distributed in both the cytoplasm and nucleus (Supplementary Fig. 10a). Furthermore, the WT MKK9–GFP was translocated into the nucleus in response to ACC in WT but not in etr1 protoplasts (Fig. 5d), even though total MKK9–GFP protein levels were similar in WT and etr1 (Supplementary Fig. 10b). Significantly, the immunoprecipitated MPK3 or MPK6 activated by MKK9a in protoplasts could directly phosphorylate EIN3 in vitro (Fig. 5e). These results suggest that direct protein phosphorylation by MKK9–MPK3/6 in the nucleus may be a key step for EIN3 protein stabilization and ethylene signalling.
a, EIN3 regulation in seedlings. Tubulin (control). b, EIN3 phosphorylated in vivo. The EIN3 protein mobility shifts without (-) or with (+) phosphatase (CIAP). c, MKK9a–GFP is localized in the nucleus. Nuclear RFP marker36 (CSRFP). MKK4a–GFP is localized in both the cytoplasm and nucleus as GFP (cont). d, MKK9–GFP is translocated into the nucleus by ACC treatment in WT but not in etr1. e, MPK3 and MPK6 directly phosphorylate T174 in a truncated EIN3 protein in vitro. tEIN3 (ref. 15), amino acids 141–352. MPK3/6 (anti-HA) and tEIN3 (Coomassie blue staining) protein levels are shown. f, The EIN3AA mutant shows enhanced protein degradation. RBC as control. g, Opposite effects of two phosphorylation sites on EIN3 stability. h, Enhanced EIN3T592A stability requires MKK9. i, T174 in EIN3 is phosphorylated in vivo by MKK9a-activated MPK6. Immunoprecipitated EIN3 was examined with a p-Thr-Pro-specific antibody. j, Active CTR1a promotes EIN3 degradation through T592 phosphorylation. Inactive InCTR1 is a control.
To map the MAPK phosphorylation site(s) in EIN3 directly, we combined computational analyses and targeted mutagenesis, which were followed by comprehensive cellular and transgenic analyses in vivo. Two robust and comprehensive motif search algorithms, Eukaryotic Linear Motif (ELM)45 and Scansite 2.0 (ref. 46), were used to search for putative MAPK phosphorylation sites and the conserved docking sites for MAPK in EIN3. There were two predictable MAPK phosphorylation sites (174T175P and 592T593P), each coupled with a putative conserved-docking motif nearby, conserved in EIN3 and EIL1 (refs 14, 19). Remarkably, the predicted MAPK conserved-docking motif (amino acids 244–252) near 174T was mutated in the ein3-3 mutant (K244D) that diminished EIN3 function10. In an in vitro assay, the T174A mutation prevented EIN3 phosphorylation by activated MPK3/6 (Fig. 5e).
To determine the in vivo function of T174 and T592 in ethylene signalling, we generated the EIN3T174A/T592A (EIN3AA) mutant, and the stability of EIN3 and EIN3AA proteins was monitored after cycloheximide (CHX) treatment in the transfected ein3 protoplasts. The non-phosphorylatable EIN3AA mutant always degraded faster than the WT EIN3 protein (Fig. 5f). Unexpectedly, when the individual phosphorylation site mutants (EIN3T174A or EIN3T592A) were tested with the same assay, we discovered their opposite effects on EIN3 stability. Compared with EIN3 (Fig. 5f), the T174A mutation enhanced EIN3 degradation whereas the T592A mutation enhanced EIN3 stability (Fig. 5g). It was clear that the endogenous MKK9 was required to stabilize EIN3T592A (Fig. 5h) in WT protoplasts with some level of constitutive ethylene signalling (Fig. 2b) that could phosphorylate T174. We also showed that the activated MPK6 phosphorylated EIN3T592A but not EIN3T174A in vivo by a specific antibody that recognized a phospho-Thr-Pro motif in protoplasts (Fig. 5i). Finally, CTR1a abolished EIN3 but not EIN3T592A, supporting a direct link between T592 phosphorylation and degradation by the CTR1 pathway (Fig. 5j). As CTR1–GFP did not accumulate in the nucleus, the results indicated that the CTR1 pathway probably acted through MAPKs (Fig. 1a) to phosphorylate T592 and promotes EIN3 degradation in the nucleus. We proposed a model that the MKK9 cascade phosphorylates T174 to promote EIN3 stability, whereas T592 is phosphorylated by an MAPK pathway mediated by CTR1 to promote EIN3 degradation.
To test our model in plants further, we generated and examined ein3 transgenic plants complemented with WT EIN3, EIN3AA, EIN3T174A and EIN3T592A. For informative comparisons and to avoid the dominant effect of EIN3 overexpression10, we selected multiple transgenic lines for each construct with similar levels of EIN3 expression as the endogenous EIN3 in WT (Supplementary Fig. 11). Consistent with the results obtained using the cellular assays, EIN3AA and EIN3T174A transgenic lines were insensitive to saturating ACC (Fig. 6a). On the contrary, the EIN3T592A lines exhibited ACC hypersensitivity. EIN3 protein accumulation patterns reflected the transgenic phenotypes (Fig. 6a). Detailed quantification of ACC dose responses confirmed that the transgenic EIN3T174A lines were more insensitive to ACC and the transgenic EIN3T592A lines were hypersensitive to ACC (Fig. 6b). The fact that the EIN3T592A lines only displayed weak constitutive ethylene phenotypes in the absence of ACC strongly supports the model that EIN3 phosphorylation on T174 by MKK9–MPK3/6 is activated by ethylene signalling and is critical for its stabilization even in the absence of T592 phosphorylation, which enhanced EIN3 degradation (Fig. 5g, j). Both the inhibition of CTR1 and activation of MKK9 are required for ethylene signalling specificity. The data explain why mkk9 exhibited moderate ethylene-insensitive phenotypes (Fig. 3) and escaped classical mutant screens, as only one of the bifurcate pathways (Fig. 6c) was inactive. The stronger constitutive ethylene signalling phenotypes in ctr1 are partly attributed to the activation or de-repression of the MKK9–MPK3/6 cascade revealed in ctr1 protoplasts (Figs 1 and 2).
a, Ethylene sensitivity assay. EIN3 protein accumulation in the transgenic lines is shown. Histone (control). b, Quantitative analyses of hypocotyl elongation of transgenic lines. Error bar, s.d. (n = 20). Asterisks indicate differences between ein3 and transgenic ein3 plants with statistical significance at *P < 0.05, **P < 0.01 and ***P < 0.001 (t-test). c, Model of the bifurcate MAPK cascades in ethylene signalling. The two EIN3 phosphorylation sites (P) with opposite functions are marked. Without ethylene, CTR1 directly or indirectly inactivates MKK9–MPK3/6 and probably activates downstream MAPKs to phosphorylate T592 to promote EIN3 degradation. Ethylene inactivates CTR1 for MKK9–MPK3/6 activation and T174 phosphorylation to stabilize EIN3. Arrow and blunt ends indicate positive and negative regulations, respectively. ACS, ACC synthase; MKKK, MAPKKK; C, cytoplasm; N, nucleus.
Discussion
Signalling specificity is fundamental to proper function of regulatory pathways in eukaryotic cells. A common theme in the evolutionarily conserved MAPK cascade signalling is the use of the same components in different signal transduction pathways. Many strategies can maximize the functions of a limited set of MAPK cascade modules for different biological responses and processes, including different partners in distinct cell types, subcellular compartmentalization, response amplitude and duration, temporal separation, and the use of scaffolding proteins22,25,26,27. In Arabidopsis, the confounding effects of MAPKs on both ethylene synthesis and signalling, as well as other stress and defence responses and diverse developmental processes, impose great challenges in defining MAPK signalling specificity22,23,24,28,29,30,31,32,33,34,35,36. We provide compelling evidence for the identification of a novel MKK9–MPK3/6 cascade that phosphorylates and stabilizes EIN3 in ethylene signalling. The MKK9–MPK3/6 cascade targets EIN3 in the nucleus to distinguish temporarily and spatially its positive ethylene signalling function from the MKK4/5–MPK6 activity on ACS6 for ethylene biosynthesis in the cytoplasm33. Further biochemical and genetic analyses reveal the surprising opposite functions of the dual EIN3 phosphorylation sites, T174 for stabilization and T592 for degradation. The simultaneous activation of the MKK9 cascade and the inhibition of the CTR1 pathway specify quantitative control of EIN3 levels and EIN3-mediated transcription. CTR1 as a unique MAPKKK appears to control bifurcate and antagonistic MAPK cascades targeted to the same key nuclear transcription factor in ethylene signalling (Fig. 6c). The flexible and quantitative mechanism can facilitate the molecular connections in a complex signal network modulated by hormonal, metabolic and environmental signals to serve an adjustable and adaptive lifestyle characteristic of plants. The involvement of CTR1 in auxin, gibberellin and glucose responses15,47 suggests that the CTR1 pathway may integrate other signals, and can be involved in phosphorylation and control of other transcription factors in distinct signalling pathways without the MKK9 partnership. It is also possible that the MKK9–MPK3/6 cascade can be modulated by signals other than ethylene and can participate in multiple stress and defence responses independent of CTR1 (Fig. 6c). Similar systematic and integrated approaches used in this study may be applicable for the identification of the subtle and redundant MAPK cascade components presumably acting downstream of CTR1. How CTR1 regulates EIN2 and inhibits MKK9–MPK3/6, whether other MAPKKKs activate MKK9, and whether EBF1 and EBF2 are also regulated by MAPK cascades, remain to be determined (Fig. 6c). There are over 100 putative MAPK cascade genes in Arabidopsis, and many are conserved in agriculturally important plants24,48. It will be interesting to elucidate how the positive and negative functions of MAPK cascades interact in the signalling networks modulated by a myriad of internal and external signals to govern essential biological processes22,23,24,25,26,27.
Methods Summary
Protoplast transient assays
Transient expression and MAPK activity assays were performed in Arabidopsis mesophyll protoplasts as described15,35,36,49. Subcellular localizations of MKK4a–GFP, MKK9a–GFP and ACC-treated MKK9–GFP were observed in transfected protoplasts by fluorescent microscopy15,36,49. Protein stability assays were conducted using WT and mutant EIN3 proteins as described15. Transfected ein3 protoplasts were incubated for 4 h before treatment with 10 µM CHX to stop de novo protein synthesis. EIN3 protein degradation was visualized by immunoblot analysis.
MPK and MKK screens
The MKK9–MPK3/6 module was identified by sensitized screens with epitope-tagged MPKs and MKKs by using immunocomplex MAPK activity35,36 and the ethylene-specific EBS-LUC reporter15 assays in ctr1 protoplasts8,49.
The mkk mutant screens
The loss-of-function mkk mutants were identified by standard methods40,41 and screened for the ethylene-specific hypocotyl response using etiolated seedlings free of other stresses10,14.
Gene expression analysis
Quantitative polymerase chain reaction with reverse transcription (qRT–PCR) was used with gene-specific primers for ERF1 and ERF5 in WT, mutants and transgenic lines.
Transgenic plants
The mkk9-1 mutant was complemented with a WT MKK9 genomic construct. The gain-of-function MKK9a transgenic lines were generated in WT, etr1-1 and ein2-1 background and used for epistasis analyses. The WT and various EIN3 mutants were introduced into ein3 plants. Multiple transgenic lines were selected with equal transgene expression for analyses of ethylene response.
EIN3 in vivo phosphorylation
We examined EIN3 accumulation and phosphorylation induced by ACC (100 µM) with etiolated seedlings in the absence or presence of an MKK inhibitor (U0126, 10 µM), a proteasome inhibitor (MG132, 50 µM) and/or a general phosphatase (calf intestine alkaline phosphatase) using protein blot analyses with a specific EIN3 antibody15. We detected in vivo EIN3 phosphorylation by activated MPK6 in protoplasts with protein blot analysis using a p-Thr-Pro-specific antibody and the EIN3 phosphorylation mutants EIN3T174A and EIN3T592A.
Online Methods
Plasmid constructs
The reporter constructs, EBS-LUC (EBS)15, WRKY29-LUC (W29)36, RD29A-LUC (R29A)35 and GH3-LUC (GH3)34, have been described previously. The 3.9 kilobases (kb) of the ERF5 (At5g47230) promoter were amplified by PCR and fused to the LUC gene to create the ERF5-LUC reporter construct. All effector constructs were generated by inserting the complementary DNA (cDNA) between the 35SC4PPDK promoter and the NOS terminator in a plant expression vector for protoplast transient assays15,34,35,36,49 or a mini-binary vector pCB302 or pBIN19 for transgenic plant analysis15. All inserts were verified by DNA sequencing.
Effector constructs
The coding regions of MPKs, MKKs and MKKKs were tagged at carboxy (C) termini with double HA, double MYC and single Flag epitopes, respectively.
The effectors used in the experiment were MPK1 (At1g10210), MPK2 (At1g59580), MPK3 (At3g45640), MPK4 (At4g01370), MPK5 (At4g11330), MPK6 (At2g43790), MPK7 (At2g18170), MPK9 (At3g18040), MPK10 (At3g59790), MPK11 (At1g01560), MPK12 (At2g46070), MPK13 (At1g07880), MPK14 (At4g36450), MKK1 (At4g26070), MKK2 (At4g29810), MKK3 (At5g40440), MKK4 (At1g51660), MKK5 (At3g21220), MKK6 (At5g56580), MKK7 (At1g18350) and MKK9 (At1g73500)24. Constitutively active forms of MKKs were generated by changing T or S in the activation domain of [T/S]XXXXX[S/T] to D or E by PCR-based site-specific mutagenesis36. MKK1a (T218E, S224D), MKK2a (T220D, T226E), MKK3a (S235E, T241D), MKK4a (T224D, S230E), MKK5a (T202E, S208E), MKK6a (S221D, T227E), MKK7a (S193E, S199D) and MKK9a (S195E, S201E) were used in the experiments. The active CTR1a and inactive InCTR1 constructs (At5g03730) are the same as previously described15,36. The EIN3 WT and mutant (T174A, T592A, and T174A and T592A as AA) constructs were generated similarly.
Arabidopsis mesophyll protoplast transient expression assay
Protoplast isolation and transient expression assays were performed as described15,34,35,36,49. For ethylene treatment, protoplast samples in culture dishes were placed in air-tight transparent containers with or without 10 p.p.m. ethylene (Alltech) for 6 h. Protoplasts were incubated in a minimal volume of medium (0.6 mm in depth) to facilitate ethylene gas perception. All protoplast transient assays were performed with UBQ10-GUS as an internal control15,34,35,36,49. All reporter activities were calculated based on LUC/GUS ratio and normalized to the values obtained without the treatment or effector expression. Subcellular localizations of activated MKKs were determined by microscopic observation of protoplasts co-transfected with MKK4a–GFP, MKK9a–GFP or GFP (Control) with a nuclear RFP marker36. Protoplast experiments were repeated at least three independent times with consistent results and presented as means with standard deviations. Protein expression was examined by immunoblot analysis using commercially available monoclonal antibodies to HA (Roche), MYC (Roche), GFP (Roche), Flag (Sigma), tubulin (Sigma), MPK3 (Sigma), histone H1 (Upstate) and phospho-threonine-proline (Cell Signaling), a polyclonal antibody to EIN3 (anti-EIN3)15, RBC (anti-RBC)15 or MPK6 (anti-MPK6) generated against the synthetic peptide LIYREALAFNPEYQ.
Protein kinase assays
MAPK in-gel kinase and immunocomplex MAPK assays were performed with myelin basic protein as a general substrate as described34,35,36. Epitope-tagged MAPKs were immunoprecipitated from lysates of the transfected protoplasts with the corresponding antibodies and analysed with [γ-32P]ATP and myelin basic protein34,35,36. Experiments were repeated at least three times with consistent results. The expression of CTR1a was not sufficient to reveal overt activation of co-expressed MAPKs in WT protoplasts, and ctr1 protoplasts were used for MAPK screens. The MAPK repression by CTR1a required its protein kinase activity, as the kinase-dead inactive InCTR1K579M (ref. 15) failed to show similar activity when expressed at the same protein level. MAPK assays were repeated at least five independent times with consistent results, and representative results are presented. As shown previously30,31, we confirmed that the same ACC treatment did not activate any endogenous MAPKs in the dominant ethylene receptor etr1-1 mutant with little ethylene perception (data not shown).
RNA isolation and transcript measurement
Total RNA was isolated by the Trizol method (Invitrogen) and 1 μg of total RNA was used for cDNA synthesis. Quantitative PCR was performed with iQ SYBR Green dye-added PCR mix according to the manufacturer’s instruction (Bio-Rad). Tubulin4 (At1g04820) or elongation initiation factor4a (ELF4a, At3g13920) transcript was used as an RT–PCR control with gene-specific primers. Experiments were repeated three times with consistent results. The primer sequences are provided in Supplementary Table 1. Each primer set was pre-tested by PCR for a single gene product. Microarray data of MPKs and MKKs transcript levels in adult leaves were collected by Genevestigator50.
Transgenic and mutant plant analyses
Arabidopsis transgenic plants were generated with WT (Col-0), etr1 (etr1-1), ein2 (ein2-1) and ein3 (ein3-1) by the floral dipping method as described before15. Transgene expression levels were measured by RT–PCR using gene-specific forward primer (MKK9_f) and a transgene specific reverse primer (Ts_r). Multiple independent transgenic lines were generated and analysed for consistency. We analysed the phenotypes of etiolated seedlings with at least two independent lines of T2 or T3 generations. The mpk3, mpk6 and mkk mutants were identified from the Salk and Syngenta SAIL T-DNA insertion collections40,41 (Supplementary Figs 3–5). The primer sequences are provided in Supplementary Table 1. The growth response assays of etiolated seedlings were performed with silver ion (Sigma) or ACC (Sigma) in the complete dark for three or four days. Seeds were germinated on MS agar (1% sucrose) media after four days of cold treatment to ensure uniform germination17. For glucose repression assays, seedlings were grown on 4% glucose (Glc) MS medium for five days under constant light (60 μmol m-2 s-1)15. For salt sensitivity assays, seedlings were grown on 100 mM NaCl 0.5 × MS medium for 15 days under constant light (60 μmol m-2 s-1). Total chlorophyll assay was performed with detached mature green leaves (28 days) treated with 1 µM ACC for three days in the dark as described10. To observe ethylene-induced changes of rosette size, plants were grown for 14 days and then transferred to chambers without or with a daily dose of 5 p.p.m. C2H4 for a week. Transcript levels of immediate early genes were measured by qRT–PCR with RNA extracted from leaves treated with 5 p.p.m. C2H4 for 1 h. Molecular and phenotypic analyses were repeated at least three times with similar results.
References
Neljubov, D. N. Uber die horizontale Nutation der Stengel von Pisum sativum und einiger anderen Pflanzen. Beih. Bot. Centralbh. 10, 129–139 (1901)
Bleecker, A. B. & Kende, H. Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18 (2000)
Chen, Y.-F., Etheridge, N. & Schaller, G. E. Ethylene signal transduction. Ann. Bot. (Lond.) 95, 901–915 (2005)
Guo, H. & Ecker, J. R. The ethylene signalling pathway: new insights. Curr. Opin. Plant Biol. 7, 40–49 (2004)
Alonso, J. M. & Stepanova, A. N. The ethylene signalling pathway. Science 306, 1513–1515 (2004)
Chang, C., Kwok, S. F., Bleecker, A. B. & Meyerowitz, E. M. Arabidopsis ethylene response gene ETR1: similarity of product to two component regulators. Science 262, 539–544 (1993)
Hua, J. & Meyerowitz, E. M. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana . Cell 94, 261–271 (1998)
Kieber, J. J., Rthenberg, M., Roman, G., Feldmann, K. A. & Ecker, J. R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441 (1993)
Alonso, J. M., Hirayama, T., Roman, G., Nourizadeh, S. & Ecker, J. R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis . Science 284, 2148–2152 (1999)
Chao, Q. et al. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ethylene-insensitive 3 and related proteins. Cell 89, 1133–1144 (1997)
Clark, K. L., Larsen, P. B., Wang, X. & Chang, C. Association of the Arabidopsis CTR1 raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl Acad. Sci. USA 95, 5401–5406 (1998)
Huang, Y., Li, H., Hutchison, C. E., Laskey, J. & Kieber, J. J. Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis . Plant J. 33, 221–233 (2003)
Solano, R., Stepanova, A., Chao, Q. & Ecker, J. R. Nuclear events in ethylene signalling: a transcriptional cascade mediated by ethylene-insensitve 3 and ethylene-response-factor 1. Genes Dev. 12, 3703–3714 (1998)
Alonso, J. M. et al. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis . Proc. Natl Acad. Sci. USA 100, 2992–2997 (2003)
Yanagisawa, S., Yoo, S.-D. & Sheen, J. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425, 521–525 (2003)
Potuschak, T. et al. EIN3-dependent regulation of plant ethylene hormone signalling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689 (2003)
Guo, H. & Ecker, J. R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677 (2003)
Gagne, J. M. et al. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl Acad. Sci. USA 101, 6803–6808 (2004)
Binder, B. M. et al. The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19, 509–523 (2007)
Drehere, K. & Callis, J. Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99, 787–822 (2007)
Smalle, J. & Vierstra, R. D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590 (2004)
Tena, G., Asai, T., Chiu, W.-L. & Sheen, J. Plant mitogen-activated protein kinase signalling cascades. Curr. Opin. Plant Biol. 4, 392–400 (2001)
Nakagami, H., Pitzschke, A. & Hirt, H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 10, 339–346 (2005)
Ichimura, K. et al. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308 (2002)
Raman, M. & Cobb, M. H. MAP kinase modules: many roads home. Curr. Biol. 13, R886–R888 (2003)
Schwartz, M. A. & Madhani, H. D. Principles of MAP kinase signalling specificity in Saccharomyces cerevisiae . Annu. Rev. Genet. 38, 725–748 (2004)
Morrison, D. K. & Davis, R. J. Regulation of MAP kinase signalling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 19, 91–118 (2003)
Takahashi, F. et al. The mitogen-activated protein kinase cascade MKK3–MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis . Plant Cell 19, 805–818 (2007)
Wang, H., Ngwenyama, N., Liu, Y., Walker, J. C. & Zhang, S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis . Plant Cell 19, 63–73 (2007)
Novikova, G. V., Moshkov, I. E., Smith, A. R. & Hall, M. A. The effect of ethylene on MAP Kinase-like activity in Arabidopsis thaliana. FEBS Lett. 474, 29–32 (2000)
Ouaked, F., Rozhon, W., Lecourieux, D. & Hirt, H. A. MAPK pathway mediates ethylene signalling in plants. EMBO J. 22, 1282–1288 (2003)
Ecker, J. R. Reentry of the ethylene MPK6 module. Plant Cell 16, 3169–3173 (2004)
Liu, Y. & Zhang, S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis . Plant Cell 16, 3386–3399 (2004)
Kovtun, Y., Chiu, W.-L., Zeng, W. & Sheen, J. Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395, 716–720 (1998)
Kovtun, Y., Chiu, W.-L., Tena, G. & Sheen, J. Function analysis of oxidative stress activated mitogen-activated protein kinase cascade in plants. Proc. Natl Acad. Sci. USA 97, 2940–2945 (2000)
Asai, T. et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983 (2002)
Wang, W., Hall, A. E., O’Malley, R. & Bleecker, A. B. Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc. Natl Acad. Sci. USA 100, 352–357 (2003)
Qu, X., Hall, B. P., Gao, Z. & Schaller, G. E. A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 7, 1–15 (2007)
Larsen, P. B. & Chang, C. The Arabidopsis eer1 mutant has enhanced ethylene responses in the hypocotyl and stem. Plant Physiol. 125, 1061–1073 (2001)
Alonso, J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003)
Sessions, A. et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994 (2002)
Cheng, W. H. et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signalling and abscisic acid biosynthesis and functions. Plant Cell 14, 2723–2743 (2002)
Cao, W.-H. et al. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 143, 707–719 (2006)
Burch-Smith, T. M., Schiff, M., Liu, Y. & Dinesh-Kumar, S. P. Efficient virus-induced gene silencing in Arabidopsis . Plant Physiol. 142, 21–27 (2006)
Puntervoll, P. et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31, 3625–3630 (2003)
Obenauer, J. C., Cantley, L. C. & Yaffe, M. B. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 (2003)
Achard, P., Vriezen, W. H., Van Der Straeten, D. & Harberd, N. P. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15, 2816–2825 (2003)
Hamel, L. P. et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11, 192–198 (2006)
Yoo, S.-D., Cho, Y.-H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572 (2007)
Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L. & Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632 (2004)
Acknowledgements
We thank the Salk Institute, Syngenta Biotechnology and A. Sessions for sharing the Arabidopsis T-DNA collections, the Arabidopsis Biological Resource Center for providing Arabidopsis mutant seeds, B. Mueller for offering quantitative PCR primers, and colleagues at Sheen Laboratory for comments on the manuscript. We appreciate S. Yanagisawa for the EBS-LUC construct, S. P. Dinesh-Kumar for sharing the VIGS constructs and protocol, and H. Guo, J. Ecker and S. Zhang for sharing unpublished data. The work was supported by grants from the National Science Foundation and the National Institutes of Health to J.S.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
The file contains Supplementary Figures S1-S11 with Legends and Supplementary Table S1. (PDF 18146 kb)
Rights and permissions
About this article
Cite this article
Yoo, SD., Cho, YH., Tena, G. et al. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451, 789–795 (2008). https://doi.org/10.1038/nature06543
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06543
This article is cited by
-
Comparative transcriptome analysis reveals the regulatory mechanisms of two tropical water lilies in response to cold stress
BMC Genomics (2023)
-
Nitric oxide alleviates salt stress through protein S-nitrosylation and transcriptional regulation in tomato seedlings
Planta (2022)
-
Recent advances in physiological and molecular mechanisms of heavy metal accumulation in plants
Environmental Science and Pollution Research (2021)
-
Role of ethylene in the regulatory mechanism underlying the abortion of ovules after fertilization in Xanthoceras sorbifolium
Plant Molecular Biology (2021)
-
The involvement of gaseous signaling molecules in plant MAPK cascades: function and signal transduction
Planta (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.