Abstract
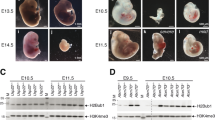
Post-translational histone modifications have important regulatory roles in chromatin structure and function1,2,3. One example of such modifications is histone ubiquitination, which occurs predominately on histone H2A and H2B. Although the recent identification of the ubiquitin ligase for histone H2A has revealed important roles for H2A ubiquitination in Hox gene silencing4,5,6 as well as in X-chromosome inactivation7,8, the enzyme(s) involved in H2A deubiquitination and the function of H2A deubiquitination are not known. Here we report the identification and functional characterization of the major deubiquitinase for histone H2A, Ubp-M (also called USP16). Ubp-M prefers nucleosomal substrates in vitro, and specifically deubiquitinates histone H2A but not H2B in vitro and in vivo. Notably, knockdown of Ubp-M in HeLa cells results in slow cell growth rates owing to defects in the mitotic phase of the cell cycle. Further studies reveal that H2A deubiquitination by Ubp-M is a prerequisite for subsequent phosphorylation of Ser 10 of H3 and chromosome segregation when cells enter mitosis. Furthermore, we demonstrate that Ubp-M regulates Hox gene expression through H2A deubiquitination and that blocking the function of Ubp-M results in defective posterior development in Xenopus laevis. This study identifies the major deubiquitinase for histone H2A and demonstrates that H2A deubiquitination is critically involved in cell cycle progression and gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nature Rev. Mol. Cell Biol. 6, 838–849 (2005)
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001)
Peterson, C. L. & Laniel, M.-A. Histones and histone modifications. Curr. Biol. 14, R546–R551 (2004)
Wang, H. et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004)
Cao, R., Tsukada, Y.-i. & Zhang, Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20, 845–854 (2005)
Wei, J., Zhai, L., Xu, J. & Wang, H. Role of Bmi1 in H2A ubiquitylation and Hox gene silencing. J. Biol. Chem. 281, 22537–22544 (2006)
Fang, J., Chen, T., Chadwick, B., Li, E. & Zhang, Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J. Biol. Chem. 279, 52812–52815 (2004)
de Napoles, M. et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663–676 (2004)
Fang, J., Wang, H. & Zhang, Y. Purification of histone methyltransferases from HeLa cells. Methods Enzymol. 377, 213–226 (2004)
Wang, H. et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8, 1207–1217 (2001)
Henry, K. W. et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17, 2648–2663 (2003)
Daniel, J. A. et al. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279, 1867–1871 (2004)
Erdjument-Bromage, H. et al. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A. 826, 167–181 (1988)
Winkler, G. et al. Isolation and mass spectrometry of transcription factor complexes. Methods 26, 260–269 (2002)
Cai, S. Y., Babbitt, R. & Marchesi, V. A mutant deubiquitinating enzyme (Ubp-M) associates with mitotic chromosomes and blocks cell division. Proc. Natl Acad. Sci. USA 96, 2828–2833 (1999)
Mimnaugh, E. G. et al. Caspase-dependent deubiquitination of monoubiquitinated nucleosomal histone H2A induced by diverse apoptogenic stimuli. Cell Death Differ. 8, 1182–1196 (2001)
Wang, H. et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22, 383–394 (2006)
Shiio, Y. & Eisenman, R. N. Histone sumoylation is associated with transcriptional repression. Proc. Natl Acad. Sci. USA 100, 13225–13230 (2003)
Robzyk, K., Recht, J. & Osley, M. A. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287, 501–504 (2000)
Henry, K. W. et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17, 2648–2663 (2003)
Emre, N. C. T. et al. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal sir2 association and gene silencing. Mol. Cell 17, 585–594 (2005)
Luger, K., Rechsteiner, T. J. & Richmond, T. J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 (1999)
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004)
Dignam, J. D., Martin, P., Shastry, B. & Roeder, R. Eukaryotic gene transcription with purified components. Methods Enzymol. 101, 582–598 (1983)
Fischle, W. et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 (2005)
Harland, R. M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36, 685–695 (1991)
Bolton, M. A. et al. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell 13, 3064–3077 (2002)
Perez-Burgos, L. et al. Generation and characterization of methyl-lysine histone antibodies. Methods Enzymol. 376, 234–254 (2003)
Song, L. et al. Ser-10 phosphorylated histone H3 is involved in cytokinesis as a chromosomal passenger. Cell Biol. Int. 31, 1184–1190 (2007)
Acknowledgements
We thank S. L. Berger and M. A. Osley for yeast strains; J. J. Hayes for XP-10 plasmid; and J. Wei for the initial cloning of Ubp-M. We also thank D. Crawford and W. S. Brooks for suggestions on cell cycle and apoptosis analysis; T. Townes and W. S. Brooks for critical reading of the manuscript; E. F. Keyser for assistance with FACS analysis; and A. Nazarian for help with mass spectrometric analysis. This work was supported by a start-up grant (to H.W.) and NCI Cancer Center Support Grant (to P.T.).
Author Contributions H.W. designed the experimental strategy, performed parts of the purification and wrote the paper; H.-Y.J. performed most of the purification, determined the substrate preference and investigated the role of Ubp-M in cell cycle progression and gene expression; L.Z. determined the substrate specificity of Ubp-M and performed the in vitro kinase reaction and nucleosome pull-down experiments; C.Y. cloned the Xenopus Hoxd10 gene and generated the RNAi-resistant Ubp-M constructs; S.N. and C.C. performed the Xenopus injection and in situ hybridization; H.E.-B. and P.T. performed mass spectrometric analysis.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1-S15 with Legends. (PDF 1263 kb)
Rights and permissions
About this article
Cite this article
Joo, HY., Zhai, L., Yang, C. et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 449, 1068–1072 (2007). https://doi.org/10.1038/nature06256
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06256
This article is cited by
-
Deubiquitinase USP16 induces gouty arthritis via Drp1-dependent mitochondrial fission and NLRP3 inflammasome activation
Arthritis Research & Therapy (2023)
-
USP16 regulates castration-resistant prostate cancer cell proliferation by deubiquitinating and stablizing c-Myc
Journal of Experimental & Clinical Cancer Research (2021)
-
Non-canonical function of DGCR8 in DNA double-strand break repair signaling and tumor radioresistance
Nature Communications (2021)
-
The deubiquitinase USP16 functions as an oncogenic factor in K-RAS-driven lung tumorigenesis
Oncogene (2021)
-
Ubiquitin signaling in cell cycle control and tumorigenesis
Cell Death & Differentiation (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.