Abstract
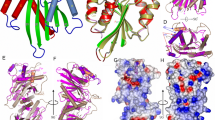
Holliday proposed a four-way DNA junction as an intermediate in homologous recombination1, and such Holliday junctions have since been identified as a central component in DNA recombination and repair2. Phage T4 endonuclease VII (endo VII) was the first enzyme shown to resolve Holliday junctions into duplex DNAs by introducing symmetrical nicks in equivalent strands3. Several Holliday junction resolvases have since been characterized4, but an atomic structure of a resolvase complex with a Holliday junction remained elusive. Here we report the crystal structure of an inactive T4 endo VII(N62D) complexed with an immobile four-way junction with alternating arm lengths of 10 and 14 base pairs. The junction is a hybrid of the conventional square-planar and stacked-X conformation. Endo VII protrudes into the junction point from the minor groove side, opening it to a 14 Å × 32 Å parallelogram. This interaction interrupts the coaxial stacking, yet every base pair surrounding the junction remains intact. Additional interactions involve the positively charged protein and DNA phosphate backbones. Each scissile phosphate that is two base pairs from the crossover interacts with a Mg2+ ion in the active site. The similar overall shape and surface charge potential of the Holliday junction resolvases endo VII, RuvC, Ydc2, Hjc and RecU, despite having different folds, active site composition and DNA sequence preference, suggest a conserved binding mode for Holliday junctions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Holliday, R. A mechanism for gene conversion in fungi. Genet. Res. 5, 282–304 (1964)
Liu, Y. & West, S. C. Happy Hollidays: 40th anniversary of the Holliday junction. Nature Rev. Mol. Cell Biol. 5, 937–944 (2004)
Mizuuchi, K., Kemper, B., Hays, J. & Weisberg, R. A. T4 endonuclease VII cleaves Holliday structures. Cell 29, 357–365 (1982)
Lilley, D. M. & White, M. F. The junction-resolving enzymes. Nature Rev. Mol. Cell Biol. 2, 433–443 (2001)
Kemper, B. & Janz, E. Function of gene 49 of bacteriophage T4. I. Isolation and biochemical characterization of very fast-sedimenting DNA. J. Virol. 18, 992–999 (1976)
Kemper, B. & Brown, D. T. Function of gene 49 of bacteriophage T4. II. Analysis of intracellular development and the structure of very fast-sedimenting DNA. J. Virol. 18, 1000–1015 (1976)
Solaro, P. C., Birkenkamp, K., Pfeiffer, P. & Kemper, B. Endonuclease VII of phage T4 triggers mismatch correction in vitro . J. Mol. Biol. 230, 868–877 (1993)
Raaijmakers, H. et al. X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J. 18, 1447–1458 (1999)
Raaijmakers, H., Tõrö, I., Birkenbihl, R., Kemper, B. & Suck, D. Conformational flexibility in T4 endonuclease VII revealed by crystallography: implications for substrate binding and cleavage. J. Mol. Biol. 308, 311–323 (2001)
Duckett, D. R. et al. The structure of the Holliday junction, and its resolution. Cell 55, 79–89 (1988)
Lilley, D. M. Structures of helical junctions in nucleic acids. Q. Rev. Biophys. 33, 109–159 (2000)
Pöhler, J. R., Giraud-Panis, M. J. & Lilley, D. M. T4 endonuclease VII selects and alters the structure of the four-way DNA junction; binding of a resolution-defective mutant enzyme. J. Mol. Biol. 260, 678–696 (1996)
Giraud-Panis, M. J. & Lilley, D. M. Near-simultaneous DNA cleavage by the subunits of the junction-resolving enzyme T4 endonuclease VII. EMBO J. 16, 2528–2534 (1997)
Giraud-Panis, M. J. & Lilley, D. M. T4 endonuclease VII. Importance of a histidine-aspartate cluster within the zinc-binding domain. J. Biol. Chem. 271, 33148–33155 (1996)
Golz, S., Christoph, A., Birkenkamp-Demtröder, K. & Kemper, B. Identification of amino acids of endonuclease VII essential for binding and cleavage of cruciform DNA. Eur. J. Biochem. 245, 573–580 (1997)
Scholz, S. R. et al. Experimental evidence for a ββα-Me-finger nuclease motif to represent the active site of the caspase-activated DNase. Biochemistry 42, 9288–9294 (2003)
Yang, W., Lee, J. Y. & Nowotny, M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006)
Stoddard, B. L. Homing endonuclease structure and function. Q. Rev. Biophys. 38, 49–95 (2005)
Khuu, P. A., Voth, A. R., Hays, F. A. & Ho, P. S. The stacked-X DNA Holliday junction and protein recognition. J. Mol. Recognit. 19, 234–242 (2006)
Hargreaves, D. et al. Crystal structure of E. coli RuvA with bound DNA Holliday junction at 6 Å resolution. Nature Struct. Biol. 5, 441–446 (1998)
Ariyoshi, M., Nishino, T., Iwasaki, H., Shinagawa, H. & Morikawa, K. Crystal structure of the Holliday junction DNA in complex with a single RuvA tetramer. Proc. Natl Acad. Sci. USA 97, 8257–8262 (2000)
Gopaul, D. N., Guo, F. & Van Duyne, G. D. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 17, 4175–4187 (1998)
Conway, A. B., Chen, Y. & Rice, P. A. Structural plasticity of the Flp–Holliday junction complex. J. Mol. Biol. 326, 425–434 (2003)
Biswas, T. et al. A structural basis for allosteric control of DNA recombination by λ integrase. Nature 435, 1059–1066 (2005)
Hadden, J. M., Déclais, A.-C., Carr, S. B., Lilley, D. M. J. & Phillips, E. V. The structural basis of Holliday junction resolution by T7 endonuclease I. Nature advance online publication. doi: 10.1038/nature06158 (2007)
Déclais, A. C., Hadden, J., Phillips, S. E. & Lilley, D. M. The active site of the junction-resolving enzyme T7 endonuclease I. J. Mol. Biol. 307, 1145–1158 (2001)
Ariyoshi, M. et al. Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli . Cell 78, 1063–1072 (1994)
Bond, C. S., Kvaratskhelia, M., Richard, D., White, M. F. & Hunter, W. N. Structure of Hjc, a Holliday junction resolvase, from Sulfolobus solfataricus . Proc. Natl Acad. Sci. USA 98, 5509–5514 (2001)
Ceschini, S. et al. Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2. EMBO J. 20, 6601–6611 (2001)
McGregor, N. et al. The structure of Bacillus subtilis RecU Holliday junction resolvase and its role in substrate selection and sequence-specific cleavage. Structure 13, 1341–1351 (2005)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
The. CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61, 458–464 (2005)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Jones, T. A., Zou, J.-Y. & Cowan, S. W. Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004)
Lavery, R. & Sklenar, H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 6, 655–667 (1989)
Luscombe, N. M., Laskowski, R. A. & Thornton, J. M. NUCPLOT: a program to generate schematic diagrams of protein–nucleic acid interactions. Nucleic Acids Res. 25, 4940–4945 (1997)
Kleywegt, G. J. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D 52, 842–857 (1996)
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991)
Acknowledgements
We thank D. Leahy and M. Gellert for critical reading of the manuscript, S. Ramon-Maiques for collecting the diffracting data of the endo VII–heteroduplex complex, and D. M. Lilley for background reading materials. C.B. thanks J. Basquin, E. Ennifar and C. Sauter for help with crystallization and data collection, and M. Nowotny and J. Y. Lee for help with manuscript preparation. This research was supported by EMBL, Deutsche Forschungsgemeinschaft and the Intramural Research Program of NIDDK, NIH.
Author Contributions C.B. carried out all experiments. The project was initiated at EMBL and finished at NIH. All authors contributed to experimental design, interpretation and manuscript preparation.
Atomic coordinates and structure factors of the endo VII–DNA complexes have been deposited in the Protein Data Bank. The accession codes are 2QNC and 2QNF for the Holliday junction and the heteroduplex complex, respectively.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Table S1 and Supplementary Figures S1-S4 with Legends. (PDF 2425 kb)
Rights and permissions
About this article
Cite this article
Biertümpfel, C., Yang, W. & Suck, D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature 449, 616–620 (2007). https://doi.org/10.1038/nature06152
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06152
This article is cited by
-
Principles of target DNA cleavage and the role of Mg2+ in the catalysis of CRISPR–Cas9
Nature Catalysis (2022)
-
Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase
Scientific Reports (2020)
-
Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1
Nature Communications (2020)
-
Junction resolving enzymes use multivalency to keep the Holliday junction dynamic
Nature Chemical Biology (2019)
-
Structural basis of sequence-specific Holliday junction cleavage by MOC1
Nature Chemical Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.