Abstract
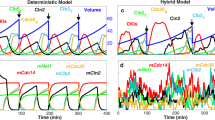
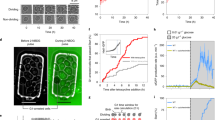
Molecular noise in gene expression can generate substantial variability in protein concentration1. However, its effect on the precision of a natural eukaryotic circuit such as the control of cell cycle remains unclear. We use single-cell imaging of fluorescently labelled budding yeast to measure times from division to budding (G1) and from budding to the next division. The variability in G1 decreases with the square root of the ploidy through a 1N/2N/4N ploidy series, consistent with simple stochastic models for molecular noise. Also, increasing the gene dosage of G1 cyclins decreases the variability in G1. A new single-cell reporter for cell protein content allows us to determine the contribution to temporal G1 variability of deterministic size control (that is, smaller cells extending G1). Cell size control contributes significantly to G1 variability in daughter cells but not in mother cells. However, even in daughters, size-independent noise is the largest quantitative contributor to G1 variability. Exit of the transcriptional repressor Whi5 from the nucleus partitions G1 into two temporally uncorrelated and functionally distinct steps. The first step, which depends on the G1 cyclin gene CLN3, corresponds to noisy size control that extends G1 in small daughters, but is of negligible duration in mothers. The second step, whose variability decreases with increasing CLN2 gene dosage, is similar in mothers and daughters. This analysis decomposes the regulatory dynamics of the Start transition into two independent modules, a size sensing module and a timing module, each of which is predominantly controlled by a different G1 cyclin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Samoilov, M. S., Price, G. & Arkin, A. P. From fluctuations to phenotypes: the physiology of noise. Sci. STKE 366, re17 (2006)
Hartwell, L. H., Culotti, J., Pringle, J. R. & Reid, B. J. Genetic control of the cell division cycle in yeast. Science 183, 46–51 (1974)
Hartwell, L. H. & Unger, M. W. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J. Cell Biol. 75, 422–435 (1977)
Johnston, G. C., Pringle, J. R. & Hartwell, L. H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105, 79–98 (1977)
Moore, S. A. Kinetic evidence for a critical rate of protein synthesis in the Saccharomyces cerevisiae yeast cell cycle. J. Biol. Chem. 263, 9674–9681 (1988)
Lord, P. G. & Wheals, A. E. Variability in individual cell cycles of Saccharomyces cerevisiae. J. Cell Sci. 50, 361–376 (1981)
Wheals, A. E. Size control-models of Saccharomyces cerevisiae cell proliferation. Mol. Cell. Biol. 2, 361–368 (1982)
Lord, P. G. & Wheals, A. E. Rate of cell cycle initiation of yeast cells when cell size is not a rate-determining factor. J. Cell Sci. 59, 183–201 (1983)
Nurse, P. Cell cycle control—both deterministic and probabilistic. Nature 286, 9–10 (1980)
Spudich, J. L. & Koshland, D. E. Non-genetic individuality: chance in the single cell. Nature 262, 467–471 (1976)
Jorgensen, P. & Tyers, M. How cells coordinate growth and division. Curr. Biol. 14, R1014–R1027 (2004)
Schroedinger, E. What is Life? (Cambridge Univ. Press, Cambridge, 1944)
Bi, E. et al. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142, 1301–1312 (1998)
Cross, F. R. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae.. Mol. Cell. Biol. 8, 4675–4684 (1988)
Nash, R., Tokiwa, G., Anand, S., Erickson, K. & Futcher, A. B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 7, 4335–4346 (1988)
Tyers, M., Tokiwa, G. & Futcher, B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12, 1955–1968 (1993)
Dirick, L., Bohm, T. & Nasmyth, K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14, 4803–4813 (1995)
Stuart, D. & Wittenberg, C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 9, 2780–2794 (1995)
Cross, F. R. Starting the cell cycle: what's the point? Curr. Opin. Cell Biol. 7, 790–797 (1995)
McInerny, C. J., Partridge, J. F., Mikesell, G. E., Creemer, D. P. & Breeden, L. L. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 11, 1277–1288 (1997)
Koch, C., Schleiffer, A., Ammerer, G. & Nasmyth, K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 10, 129–141 (1996)
Wijnen, H., Landman, A. & Futcher, B. The G(1) cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol. Cell. Biol. 22, 4402–4418 (2002)
Holstege, F. C. P. et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 (1998)
Bean, J. M., Siggia, E. D. & Cross, F. R. Coherence and timing of cell cycle start examined at single-cell resolution. Mol. Cell 21, 3–14 (2006)
Elliott, S. G. & McLaughlin, C. S. Rate of macromolecular synthesis through the cell cycle of the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 75, 4384–4388 (1978)
Donnan, L. & John, P. C. Cell cycle control by timer and sizer in Chlamydomonas. Nature 304, 630–633 (1983)
Sveiczer, A., Novak, B. & Mitchinson, J. M. The size control of fission yeast revisited. J. Cell Sci. 109, 2947–2957 (1996)
Costanzo, M. et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117, 899–913 (2004)
de Bruin, R. A. M., McDonald, W. H., Kalashnikova, T. I., Yates, J. & Wittenberg, C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117, 887–898 (2004)
Wittenberg, C. & Reed, S. I. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24, 2746–2755 (2005)
Acknowledgements
We thank J. Haber, E. Bi and the NCRR Yeast Resource Center, University of Washington, for plasmids and strains; P. Nurse and T. Ryan for discussions; and N. Buchler, B. Drapkin, J. Robbins, J. Roberts and J. Widom for comments on the manuscript. This work was supported by the National Institute of Health (J.M.S., E.D.S., F.R.C.), the Howard Hughes Medical Institute (J.M.B.), and the National Science Foundation (E.D.S.).
Author Contributions Experimental work by S.D. and J.M.S.; project planning, data analysis and manuscript preparation by all authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains the Supplementary Materials and Methods, Supplementary Discussion, Supplementary Tables S1- S12 and Supplementary Figures S1-S12. The discussion concerns the statistical analysis of the correlation of αT and ln(Mbirth), the analysis of the independence of the two regulatory steps of Start and a more detailed analysis of movies in glycerol/ethanol. The Supplementary Tables show the cell cycle timing analysis, the analysis of cell size at budding and of growth rates of individual cells. The Supplementary Figures show additional data concerning noise reduction by ploidy and gene dosage, the effect of CLN2 gene on Start and the distribution of growth rates of individual cells. (PDF 916 kb)
Rights and permissions
About this article
Cite this article
Talia, S., Skotheim, J., Bean, J. et al. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448, 947–951 (2007). https://doi.org/10.1038/nature06072
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06072
This article is cited by
-
Cell cycle-linked vacuolar pH dynamics regulate amino acid homeostasis and cell growth
Nature Metabolism (2023)
-
Temporal segregation of biosynthetic processes is responsible for metabolic oscillations during the budding yeast cell cycle
Nature Metabolism (2023)
-
Cell region fingerprints enable highly precise single-cell tracking and lineage reconstruction
Nature Methods (2022)
-
Sublinear scaling of the cellular proteome with ploidy
Nature Communications (2022)
-
A continuous-time stochastic Boolean model provides a quantitative description of the budding yeast cell cycle
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.