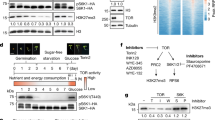
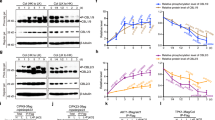
Abstract
Photosynthetic plants are the principal solar energy converter sustaining life on Earth. Despite its fundamental importance, little is known about how plants sense and adapt to darkness in the daily light–dark cycle, or how they adapt to unpredictable environmental stresses that compromise photosynthesis and respiration and deplete energy supplies. Current models emphasize diverse stress perception and signalling mechanisms1,2. Using a combination of cellular and systems screens, we show here that the evolutionarily conserved Arabidopsis thaliana protein kinases, KIN10 and KIN11 (also known as AKIN10/At3g01090 and AKIN11/At3g29160, respectively), control convergent reprogramming of transcription in response to seemingly unrelated darkness, sugar and stress conditions. Sensing and signalling deprivation of sugar and energy, KIN10 targets a remarkably broad array of genes that orchestrate transcription networks, promote catabolism and suppress anabolism. Specific bZIP transcription factors partially mediate primary KIN10 signalling. Transgenic KIN10 overexpression confers enhanced starvation tolerance and lifespan extension, and alters architecture and developmental transitions. Significantly, double kin10 kin11 deficiency abrogates the transcriptional switch in darkness and stress signalling, and impairs starch mobilization at night and growth. These studies uncover surprisingly pivotal roles of KIN10/11 in linking stress, sugar and developmental signals to globally regulate plant metabolism, energy balance, growth and survival. In contrast to the prevailing view that sucrose activates plant SnRK1s (Snf1-related protein kinases)3,4,5,6, our functional analyses of Arabidopsis KIN10/11 provide compelling evidence that SnRK1s are inactivated by sugars and share central roles with the orthologous yeast Snf1 and mammalian AMPK in energy signalling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Yamaguchi-Shinozaki, K. & Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803 (2006)
Hasegawa, P. M., Bressan, R. A., Zhu, J.-K. & Bohnert, H. J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 57, 463–499 (2000)
Halford, N. G. et al. Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J. Exp. Bot. 54, 467–475 (2003)
Bhalerao, R. P. et al. Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl Acad. Sci. USA 96, 5322–5327 (1999)
Purcell, P. C., Smith, A. M. & Halford, N. G. Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J. 14, 195–202 (1998)
Tiessen, A. et al. Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J. 35, 490–500 (2003)
Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L. & Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632 (2004)
Fujiki, Y. et al. Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant. 111, 345–352 (2001)
Moore, B. et al. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336 (2003)
Rolland, F., Baena-González, E. & Sheen, J. Sugar sensing and signaling in plants: conserved and novel Mechanisms. Annu. Rev. Plant Biol. 57, 675–709 (2006)
Lin, J. F. & Wu, S. H. Molecular events in senescing Arabidopsis leaves. Plant J. 39, 612–628 (2004)
Price, J., Laxmi, A., St Martin, S. K. & Jang, J. C. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16, 2128–2150 (2004)
Palenchar, P. M., Kouranov, A., Lejay, L. V. & Coruzzi, G. M. Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biol. 5, R91 (2004)
Hardie, D. G., Carling, D. & Carlson, M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67, 821–855 (1998)
Kahn, B. B., Alquier, T., Carling, D. & Hardie, D. G. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 1, 15–25 (2005)
Jakoby, M. et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111 (2002)
Zhang, Y. et al. Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J. 28, 431–441 (2001)
Radchuk, R., Radchuk, V., Weschke, W., Borisjuk, L. & Weber, H. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 140, 263–278 (2006)
Bläsing, O. E. et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17, 3257–3281 (2005)
Buchanan-Wollaston, V. et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42, 567–585 (2005)
Contento, A. L., Kim, S. J. & Bassham, D. C. Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135, 2330–2347 (2004)
Thimm, O. et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 (2004)
Schluepmann, H., Pellny, T., van Dijken, A., Smeekens, S. & Paul, M. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 100, 6849–6854 (2003)
Gomez, L. D., Baud, S., Gilday, A., Li, Y. & Graham, I. A. Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J. 46, 69–84 (2006)
Sugden, C., Donaghy, P. G., Halford, N. G. & Hardie, D. G. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 120, 257–274 (1999)
Kaiser, W. M. & Huber, S. C. Post-translational regulation of nitrate reductase: Mechanism, physiological relevance and environmental triggers. J. Exp. Bot. 52, 1981–1989 (2001)
Li, Y. et al. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res. 16, 414–427 (2006)
Burch-Smith, T. M., Schiff, M., Liu, Y. & Dinesh-Kumar, S. P. Efficient virus induced gene silencing in Arabidopsis thaliana. Plant Physiol. 142, 21–27 (2006)
Thelander, M., Olsson, T. & Ronne, H. Snf1-related protein kinase 1 is needed for growth in a normal day–night light cycle. EMBO J. 23, 1900–1910 (2004)
Smith, A. M., Zeeman, S. C. & Smith, S. M. Starch degradation. Annu. Rev. Plant Biol. 56, 73–98 (2005)
Kovtun, Y., Chiu, W.-L., Zeng, W. & Sheen, J. Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395, 716–720 (1998)
Kovtun, Y., Chiu, W.-L., Tena, G. & Sheen, J. Functional analysis of oxidative stress-activated MAPK cascade in plants. Proc. Natl Acad. Sci. USA 97, 2940–2945 (2000)
Hwang, I. & Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389 (2001)
Guo, Y., Halfter, U., Ishitani, M. & Zhu, J. K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13, 1383–1400 (2001)
Boudsocq, M., Barbier-Brygoo, H. & Lauriere, C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279, 41758–41766 (2004)
Lam, H. M., Peng, S. S. & Coruzzi, G. M. Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol. 106, 1347–1357 (1994)
Sheen, J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475 (2001)
Gonzali, S. et al. The use of microarrays to study the anaerobic response in Arabidopsis. Ann. Bot. (Lond.) 96, 661–668 (2005)
Loreti, E., Poggi, A., Novi, G., Alpi, A. & Perata, P. A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 137, 1130–1138 (2005)
Cheng, S.-H., Sheen, J., Gerrish, C. & Bolwell, G. P. Molecular identification of phenylalanine ammonia-lyase as a substrate of a specific constitutively active Arabidopsis CDPK expressed in maize protoplasts. FEBS Lett. 503, 185–188 (2001)
Huang, J. Z. & Huber, S. C. Phosphorylation of synthetic peptides by a CDPK and plant SNF1-related protein kinase. Influence of proline and basic amino acid residues at selected positions. Plant Cell Physiol. 42, 1079–1087 (2001)
Liu, Y., Schiff, M. & Dinesh-Kumar, S. P. Virus-induced gene silencing in tomato. Plant J. 31, 777–786 (2002)
Ryu, C. M. A. n. a. n. d. A., Kang, L. & Mysore, K. S. Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J. 40, 322–331 (2004)
Burch-Smith, T. M., Anderson, J. C., Martin, G. B. & Dinesh-Kumar, S. P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39, 734–746 (2004)
Kötting, O. et al. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 137, 242–52 (2005)
Acknowledgements
We thank S. P. Dinesh-Kumar for generously sharing the TRV-based vectors and the VIGS protocol for Arabidopsis plants before publication, O. Thimm and M. Stitt for the MAPMAN program and the original functional classification files, J.C. Jang, S. Wu and D.C. Bassham for sharing the original GeneChip data, B. Wittner for consultation on the RankProd analysis, Q. Hall for the pQH29 RNAi vector, L. Zhou for the DIN6-LUC construct, L. Shan and P. He for the VIGS GFP control vector and the infiltration procedure, J. Bush for plant management, K. Chu for data analysis, and the Sheen laboratory members for stimulating discussions. The work was supported by grants from the National Science Foundation and National Institute of Health to J.S. F.R. was supported by a return grant from the Belgian Office for Scientific, Technical and Cultural Affairs and fellowships from the Belgian American Educational Foundation and the Research Foundation–Flanders (FWO–Vlaanderen).
All microarray data are available at the Gene Expression Array Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE8248 and GSE8257.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
All microarray data are available at the Gene Expression Array Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE8248 and GSE8257. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1-S9 with Legends, Supplementary Tables S1-S7, Supplementary Methods and additional references. (PDF 7241 kb)
Supplementary Table S1
This file contains Supplementary Table S1 with global gene expression regulation by KIN10 and hypoxic conditions. Raw data. The document contains an Excel file with original unfiltered data from two biologically independent replicates of the KIN10 experiment and one hypoxia experiment (Affymetrix ATH1 Arabidopsis GeneChips) after data compilation and normalization using the GCOS v. 1.0 software. (XLS 17318 kb)
Supplementary Table S3
This file contains Supplementary Table S3 with global gene expression regulation by KIN10 (1021 genes). Data filtered as outlined in Supplementary Fig. S6). (XLS 389 kb)
Supplementary Table S4
This file contains Supplementary Table S4 with the transcriptional program induced by KIN10 markedly overlaps with that induced by starvation conditions and is antagonized by increased sugar availability (600 genes). The genes listed in Supplementary Table S3 were subjected to more stringent filtering by comparing their expression to published microarray datasets as explained in the text and outlined in Supplementary Fig. S6. (XLS 297 kb)
Supplementary Table S7
This file contains Supplementary Table S7 with sequences of primers used in this study. (XLS 28 kb)
Rights and permissions
About this article
Cite this article
Baena-González, E., Rolland, F., Thevelein, J. et al. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942 (2007). https://doi.org/10.1038/nature06069
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06069
This article is cited by
-
Contemporary understanding of transcription factor regulation of terpenoid biosynthesis in plants
Planta (2024)
-
Seagrass genomes reveal ancient polyploidy and adaptations to the marine environment
Nature Plants (2024)
-
Characterization of the basic leucine zipper transcription factor family of Neoporphyra haitanensis and its role in acclimation to dehydration stress
BMC Plant Biology (2023)
-
The Inferior Grain Filling Initiation Promotes the Source Strength of Rice Leaves
Rice (2023)
-
Transcriptomic analysis implicates ABA signaling and carbon supply in the differential outgrowth of petunia axillary buds
BMC Plant Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.