Abstract
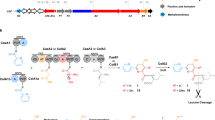
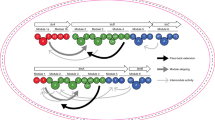
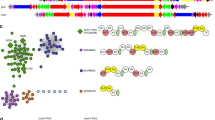
The unrelenting emergence of antibiotic-resistant bacterial pathogens demands the investigation of antibiotics with new modes of action. The pseudopeptide antibiotic andrimid is a nanomolar inhibitor of the bacterial acetyl-CoA carboxylase that catalyses the first committed step in prokaryotic fatty acid biosynthesis1. Recently, the andrimid (adm) biosynthetic gene cluster was isolated and heterologously expressed in Escherichia coli2. This establishes a heterologous biological host in which to rapidly probe features of andrimid formation and to use biosynthetic engineering to make unnatural variants of this important and promising new class of antibiotics. Bioinformatic analysis of the adm cluster revealed a dissociated biosynthetic assembly system lacking canonical amide synthases between the first three carrier protein domains. Here we report that AdmF, a transglutaminase (TGase) homologue, catalyses the formation of the first amide bond, an N-acyl-β-peptide link, in andrimid biosynthesis. Hence, AdmF is a newly discovered biosynthetic enzyme that acts as a stand-alone amide synthase between protein-bound, thiotemplated substrates in an antibiotic enzymatic assembly line. TGases (enzyme class (EC) 2.3.2.13) normally catalyse the cross-linking of (poly)peptides by creating isopeptidic bonds between the γ-carboxamide group of a glutamine side chain of one protein and various amine donors, including lysine side chains3. To the best of our knowledge, the present study constitutes the first report of a TGase-like enzyme recruited for the assembly of an antibiotic. Moreover, genome mining using the AdmF sequence yielded additional TGases in unassigned natural product biosynthetic pathways. With many more microbial genomes being sequenced, such a strategy could potentially unearth biosynthetic pathways producing new classes of antibiotics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Freiberg, C. et al. Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors with antibacterial activity. J. Biol. Chem. 279, 26066–26073 (2004)
Jin, M., Fischbach, M. A. & Clardy, J. A biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J. Am. Chem. Soc. 128, 10660–10661 (2006)
Walsh, C. T. Posttranslational modification of proteins, expanding nature's inventory (Roberts and Company Publishers, Englewood, Colorado, 2006)
Needham, J., Kelly, M. T., Ishige, M. & Andersen, R. J. Andrimid and moiramides a–c, metabolites produced in culture by a marine isolate of the bacterium Pseudomonas fluorescens: Structure elucidation and biosynthesis. J. Org. Chem. 59, 2058–2063 (1994)
Oclarit, J. M. et al. Anti-Bacillus substance in the marine sponge, Hyatella species, produced by an associated Vibrio species bacterium. Microbios 78, 7–16 (1994)
Fredenhagen, A. et al. Andrimid, a new peptide antibiotic produced by an intracellular bacterial symbiont isolated from a brown planthopper. J. Am. Chem. Soc. 109, 4409–4411 (1987)
Fischbach, M. A. & Walsh, C. T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 (2006)
Xiang, L., Moore, B. & Inactivation, S. Complementation, and heterologous expression of encP, a novel bacterial phenylalanine ammonia-lyase gene. J. Biol. Chem. 277, 32505–32509 (2002)
Christenson, S. D., Wu, W., Spies, M. A., Shen, B. & Toney, M. D. Kinetic analysis of the 4-methylideneimidazole-5-one-containing tyrosine aminomutase in enediyne antitumor antibiotic C-1027 biosynthesis. Biochemistry 42, 12708–12718 (2003)
Walker, K. D., Klettke, K., Akiyama, T. & Croteau, R. Cloning, heterologous expression, and characterization of a phenylalanine aminomutase involved in taxol biosynthesis. J. Biol. Chem. 279, 53947–53954 (2004)
Yee, V. C. et al. Three-dimensional structure of a transglutaminase: Human blood coagulation factor XIII. Proc. Natl Acad. Sci. USA 91, 7296–7300 (1994)
Keating, T. A. & Walsh, C. T. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Chem. Biol. 3, 598–606 (1999)
Liolios, K., Tavernarakis, N., Hugenholtz, P. & Kyrpides, N. C. The genomes on line database (gold) v.2: A monitor of genome projects worldwide. Nucleic Acids Res. 34, D332–D334 (2006)
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000)
Quadri, L. E. N. et al. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37, 1585–1595 (1998)
Staab, H. A., Lueking, M. & Duerr, F. H. The preparation of imidazolides. Synthesis of amides, hydrazides, and hydroxamic acids by the imidazolide method. Chem. Ber. 95, 1275–1283 (1962)
Tonge, P. J. et al. Localized electron polarization in a substrate analog binding to the active site of enoyl-CoA hydratase: Raman spectroscopic and conformational analyses of rotamers of hexadienoyl thiolesters. Biospectroscopy 1, 387–394 (1995)
Sampson, B. A., Misra, R. & Benson, S. A. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 122, 491–501 (1989)
Molnar-Perl, I. Derivatization and chromatographic behavior of the o-phthaldialdehyde amino acid derivatives obtained with various SH-group-containing additives. J. Chromatogr. A. 913, 283–302 (2001)
Acknowledgements
We thank J. Clardy for providing cosmid 2194C1 and M. Fischbach for helpful discussions. This work was supported in part by the NIH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-10 with Legends and Supplementary Tables 1-3. (PDF 755 kb)
Rights and permissions
About this article
Cite this article
Fortin, P., Walsh, C. & Magarvey, N. A transglutaminase homologue as a condensation catalyst in antibiotic assembly lines. Nature 448, 824–827 (2007). https://doi.org/10.1038/nature06068
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06068
This article is cited by
-
Fine-tuning gene expression of regulator AdmX for improved biosynthesis of andrimid in Erwinia persicina BST187
Applied Microbiology and Biotechnology (2023)
-
Amino acid (acyl carrier protein) ligase-associated biosynthetic gene clusters reveal unexplored biosynthetic potential
Molecular Genetics and Genomics (2023)
-
The hidden enzymology of bacterial natural product biosynthesis
Nature Reviews Chemistry (2019)
-
Computational redesign of enzymes for regio- and enantioselective hydroamination
Nature Chemical Biology (2018)
-
Autonomous bacterial localization and gene expression based on nearby cell receptor density
Molecular Systems Biology (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.