Abstract
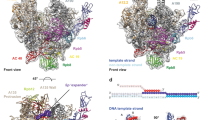
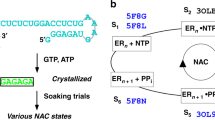
The mechanism of substrate loading in multisubunit RNA polymerase is crucial for understanding the general principles of transcription yet remains hotly debated. Here we report the 3.0-Å resolution structures of the Thermus thermophilus elongation complex (EC) with a non-hydrolysable substrate analogue, adenosine-5′-[(α,β)-methyleno]-triphosphate (AMPcPP), and with AMPcPP plus the inhibitor streptolydigin. In the EC/AMPcPP structure, the substrate binds to the active (‘insertion’) site closed through refolding of the trigger loop (TL) into two α-helices. In contrast, the EC/AMPcPP/streptolydigin structure reveals an inactive (‘preinsertion’) substrate configuration stabilized by streptolydigin-induced displacement of the TL. Our structural and biochemical data suggest that refolding of the TL is vital for catalysis and have three main implications. First, despite differences in the details, the two-step preinsertion/insertion mechanism of substrate loading may be universal for all RNA polymerases. Second, freezing of the preinsertion state is an attractive target for the design of novel antibiotics. Last, the TL emerges as a prominent target whose refolding can be modulated by regulatory factors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Temiakov, D. et al. Structural basis for substrate selection by t7 RNA polymerase. Cell 116, 381–391 (2004)
Yin, Y. W. & Steitz, T. A. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116, 393–404 (2004)
Landick, R. NTP-entry routes in multi-subunit RNA polymerases. Trends Biochem. Sci. 30, 651–654 (2005)
Svetlov, V., Vassylyev, D. G. & Artsimovitch, I. Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J. Biol. Chem. 279, 38087–38090 (2004)
Kettenberger, H., Armache, K. J. & Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16, 955–965 (2004)
Westover, K. D., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell 119, 481–489 (2004)
Sosunov, V. et al. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 22, 2234–2244 (2003)
Foster, J. E., Holmes, S. F. & Erie, D. A. Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell 106, 243–252 (2001)
Gong, X. Q., Zhang, C., Feig, M. & Burton, Z. F. Dynamic error correction and regulation of downstream bubble opening by human RNA polymerase II. Mol. Cell 18, 461–470 (2005)
Vassylyev, D. G., Vassylyeva, M. N., Perederina, A., Tahirov, T. H. & Artsimovitch, I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature doi: 10.1038/nature05932 (this issue)
Zhang, G. et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 98, 811–824 (1999)
Nickels, B. E. & Hochschild, A. Regulation of RNA polymerase through the secondary channel. Cell 118, 281–284 (2004)
Wang, D., Bushnell, D. A., Westover, K. D., Kaplan, C. D. & Kornberg, R. D. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127, 941–954 (2006)
Epshtein, V. et al. Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol. Cell 10, 623–634 (2002)
Gnatt, A. L., Cramer, P., Fu, J., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292, 1876–1882 (2001)
Bar-Nahum, G. et al. A ratchet mechanism of transcription elongation and its control. Cell 120, 183–193 (2005)
Doublie, S., Tabor, S., Long, A. M., Richardson, C. C. & Ellenberger, T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391, 251–258 (1998)
Artsimovitch, I. et al. Structural basis for transcription regulation by alarmone ppGpp. Cell 117, 299–310 (2004)
Dieci, G. et al. A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J. 14, 3766–3776 (1995)
Sosunov, V. et al. The involvement of the aspartate triad of the active center in all catalytic activities of multisubunit RNA polymerase. Nucleic Acids Res. 33, 4202–4211 (2005)
Zaychikov, E. et al. Mapping of catalytic residues in the RNA polymerase active center. Science 273, 107–109 (1996)
Langelier, M. F. et al. The highly conserved glutamic acid 791 of Rpb2 is involved in the binding of NTP and Mg(B) in the active center of human RNA polymerase II. Nucleic Acids Res. 33, 2629–2639 (2005)
Lee, J., Kashlev, M., Borukhov, S. & Goldfarb, A. A β subunit mutation disrupting the catalytic function of Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA 88, 6018–6022 (1991)
Werner, F. & Weinzierl, R. O. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell 10, 635–646 (2002)
Holmes, S. F., Santangelo, T. J., Cunningham, C. K., Roberts, J. W. & Erie, D. A. Kinetic investigation of Escherichia coli RNA polymerase mutants that influence nucleotide discrimination and transcription fidelity. J. Biol. Chem. 281, 18677–18683 (2006)
Temiakov, D. et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol. Cell 19, 655–666 (2005)
Tuske, S. et al. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell 122, 541–552 (2005)
Artsimovitch, I., Svetlov, V., Murakami, K. S. & Landick, R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J. Biol. Chem. 278, 12344–12355 (2003)
Vassylyev, D. G. et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417, 712–719 (2002)
Abbondanzieri, E. A., Greenleaf, W. J., Shaevitz, J. W., Landick, R. & Block, S. M. Direct observation of base-pair stepping by RNA polymerase. Nature 438, 460–465 (2005)
Huang, J., Brieba, L. G. & Sousa, R. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry 39, 11571–11580 (2000)
Laptenko, O., Lee, J., Lomakin, I. & Borukhov, S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 22, 6322–6334 (2003)
Sosunova, E. et al. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc. Natl Acad. Sci. USA 100, 15469–15474 (2003)
Toulokhonov, I., Zhang, J., Palangat, M. & Landick, R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell (in the press)
Kashkina, E. et al. Elongation complexes of Thermus thermophilus RNA polymerase that possess distinct translocation conformations. Nucleic Acids Res. 34, 4036–4045 (2006)
Navaza, J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 57, 1367–1372 (2001)
Brunger, A. T. et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24, 946–950 (1991)
Esnouf, R. M. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 55, 938–940 (1999)
Merrit, E. A. & Bacon, D. J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997)
Sidorenkov, I., Komissarova, N. & Kashlev, M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol. Cell 2, 55–64 (1998)
Acknowledgements
We thank V. Svetlov for help with sequence alignments and critical reading of the manuscript. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Energy Research. This work was supported by grants from the NIH to D.G.V., I.A. and R.L.
Author Contributions D.G.V. determined and analysed the structure and supervised the project. M.N.V. performed crystallization and data collection. J.Z. and M.P. performed biochemical analysis of the trigger loop mutants under the guidance of R.L. I.A. performed biochemical analysis of the T. thermophilus TECs. D.G.V., I.A. and R.L. jointly wrote the manuscript.
The atomic coordinates and structure factors of the ttEC/AMPcPP and ttEC/AMPcPP/Stl complexes are deposited in the Protein Data Bank under accession numbers 2O5J and 2PPB, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The atomic coordinates and structure factors of the ttEC/AMPcPP and ttEC/AMPcPP/Stl complexes are deposited in the Protein Data Bank under accession numbers 2O5J and 2PPB, respectively. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Discussion divided into two sections: ‘Pre-insertion state as an intermediate in the nucleotide addition cycle’ and ‘Comparison of the bacterial and eukaryotic substrate complexes’; Supplementary Tables 1-2 and Supplementary Figures 1-15 with Legends (PDF 2272 kb)
Rights and permissions
About this article
Cite this article
Vassylyev, D., Vassylyeva, M., Zhang, J. et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature 448, 163–168 (2007). https://doi.org/10.1038/nature05931
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05931
This article is cited by
-
Tail-tape-fused virion and non-virion RNA polymerases of a thermophilic virus with an extremely long tail
Nature Communications (2024)
-
Evolutionary conservation of the fidelity of transcription
Nature Communications (2023)
-
A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase
Nature Communications (2023)
-
Structural basis of transcription recognition of a hydrophobic unnatural base pair by T7 RNA polymerase
Nature Communications (2023)
-
Structural basis of R-loop recognition by the S9.6 monoclonal antibody
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.