Abstract
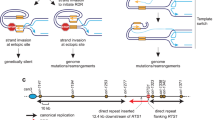
DNA double-strand breaks (DSBs) are potentially lethal lesions that arise spontaneously during normal cellular metabolism, as a consequence of environmental genotoxins or radiation, or during programmed recombination processes. Repair of DSBs by homologous recombination generally occurs by gene conversion resulting from transfer of information from an intact donor duplex to both ends of the break site of the broken chromosome1. In mitotic cells, gene conversion is rarely associated with reciprocal exchange and thus limits loss of heterozygosity for markers downstream of the site of repair and restricts potentially deleterious chromosome rearrangements2,3,4,5. DSBs that arise by replication fork collapse or by erosion of uncapped telomeres have only one free end and are thought to repair by strand invasion into a homologous duplex DNA followed by replication to the chromosome end (break-induced replication, BIR)6. BIR from one of the two ends of a DSB would result in loss of heterozygosity, suggesting that BIR is suppressed when DSBs have two ends so that repair occurs by the more conservative gene conversion mechanism. Here we show that BIR can occur by several rounds of strand invasion, DNA synthesis and dissociation. We further show that chromosome rearrangements can occur during BIR if dissociation and reinvasion occur within dispersed repeated sequences. This dynamic process could function to promote gene conversion by capture of the displaced invading strand at two-ended DSBs to prevent BIR.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Symington, L. S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630–670 (2002)
Jinks-Robertson, S. & Petes, T. D. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics 114, 731–752 (1986)
Malkova, A., Ivanov, E. L. & Haber, J. E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl Acad. Sci. USA 93, 7131–7136 (1996)
Nickoloff, J. A., Sweetser, D. B., Clikeman, J. A., Khalsa, G. J. & Wheeler, S. L. Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics 153, 665–679 (1999)
Stark, J. M. & Jasin, M. Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol. Cell. Biol. 23, 733–743 (2003)
McEachern, M. J. & Haber, J. E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75, 111–135 (2006)
Morrow, D. M., Connelly, C. & Hieter, P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147, 371–382 (1997)
Davis, A. P. & Symington, L. S. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 24, 2344–2351 (2004)
Bartsch, S., Kang, L. E. & Symington, L. S. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 20, 1194–1205 (2000)
Aguilera, A. Double-strand break repair: are Rad51/RecA–DNA joints barriers to DNA replication? Trends Genet. 17, 318–321 (2001)
Ferguson, D. O. & Holloman, W. K. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc. Natl Acad. Sci. USA 93, 5419–5424 (1996)
Nassif, N., Penney, J., Pal, S., Engels, W. R. & Gloor, G. B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14, 1613–1625 (1994)
Kraus, E., Leung, W. Y. & Haber, J. E. Break-induced replication: a review and an example in budding yeast. Proc. Natl Acad. Sci. USA 98, 8255–8262 (2001)
Adams, M. D., McVey, M. & Sekelsky, J. J. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299, 265–267 (2003)
McVey, M., Adams, M., Staeva-Vieira, E. & Sekelsky, J. J. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167, 699–705 (2004)
Merker, J. D., Dominska, M. & Petes, T. D. Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 165, 47–63 (2003)
White, M. A. & Petes, T. D. Analysis of meiotic recombination events near a recombination hotspot in the yeast Saccharomyces cerevisiae. Curr. Genet. 26, 21–30 (1994)
Dunham, M. J. et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 99, 16144–16149 (2002)
Boeke, J. D., Lacroute, F. & Fink, G. R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197, 345–346 (1984)
Koszul, R., Caburet, S., Dujon, B. & Fischer, G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 23, 234–243 (2004)
Lemoine, F. J., Degtyareva, N. P., Lobachev, K. & Petes, T. D. Chromosomal translocations in yeast induced by low levels of DNA polymerase: a model for chromosome fragile sites. Cell 120, 587–598 (2005)
Malkova, A., Naylor, M. L., Yamaguchi, M., Ira, G. & Haber, J. E. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol. Cell. Biol. 25, 933–944 (2005)
Ray, A., Machin, N. & Stahl, F. W. A. DNA double chain break stimulates triparental recombination in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 86, 6225–6229 (1989)
Schmidt, K. H., Wu, J. & Kolodner, R. D. Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae homolog of the Bloom's syndrome protein. Mol. Cell. Biol. 26, 5406–5420 (2006)
Schwartz, D. C. & Cantor, C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37, 67–75 (1984)
Lemoine, S., Combes, F., Servant, N. & Le Crom, S. Goulphar: rapid access and expertise for standard two-color microarray normalization methods. BMC Bioinformatics 7, 467 (2006)
Acknowledgements
We thank W. K. Holloman and members of the Symington and Dujon laboratories for comments on the manuscript, and T. Petes for providing the haploid strains with restriction site polymorphisms. These studies were supported by grants from the NIH, and by a postdoctoral fellowship from La Ligue Contre Le Cancer to B.L.
Author Contributions C.E.S. characterized template switching between CFV1 or CFV2 and the marked chromosome III homologues. B.L. designed and performed the experiments and analysed the data characterizing chromosome translocations by comparative genome hybridization. L.S.S. designed the experiments, analysed the data and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figure S1 with Legend and additional references. (PDF 179 kb)
Rights and permissions
About this article
Cite this article
Smith, C., Llorente, B. & Symington, L. Template switching during break-induced replication. Nature 447, 102–105 (2007). https://doi.org/10.1038/nature05723
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05723
This article is cited by
-
Break-induced replication orchestrates resection-dependent template switching
Nature (2023)
-
Pathway choice in the alternative telomere lengthening in neoplasia is dictated by replication fork processing mediated by EXD2’s nuclease activity
Nature Communications (2023)
-
Tracking break-induced replication shows that it stalls at roadblocks
Nature (2021)
-
Delayed DNA break repair for genome stability
Nature Cell Biology (2021)
-
Complete sequences of Schizosaccharomyces pombe subtelomeres reveal multiple patterns of genome variation
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.