Abstract
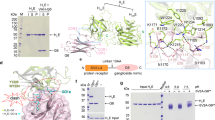
Botulinum neurotoxins (BoNTs) are potent bacterial toxins that cause paralysis at femtomolar concentrations1 by blocking neurotransmitter release. A ‘double receptor’ model has been proposed in which BoNTs recognize nerve terminals via interactions with both gangliosides and protein receptors that mediate their entry2. Of seven BoNTs (subtypes A–G), the putative receptors for BoNT/A3,4, BoNT/B5,6 and BoNT/G7 have been identified, but the molecular details that govern recognition remain undefined. Here we report the crystal structure of full-length BoNT/B in complex with the synaptotagmin II (Syt-II) recognition domain at 2.6 Å resolution. The structure of the complex reveals that Syt-II forms a short helix that binds to a hydrophobic groove within the binding domain of BoNT/B. In addition, mutagenesis of amino acid residues within this interface on Syt-II affects binding of BoNT/B. Structural and sequence analysis reveals that this hydrophobic groove is conserved in the BoNT/G and BoNT/B subtypes, but varies in other clostridial neurotoxins. Furthermore, molecular docking studies using the ganglioside GT1b indicate that its binding site is more extensive than previously proposed and might form contacts with both BoNT/B and synaptotagmin. The results provide structural insights into how BoNTs recognize protein receptors and reveal a promising target for blocking toxin–receptor recognition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Simpson, L. L. Identification of the characteristics that underlie botulinum toxin potency: implications for designing novel drugs. Biochimie 82, 943–953 (2000)
Montecucco, C. How do tetanus and botulinum toxins bind to neuronal membranes?. Trends Biochem. Sci. 11, 315–317 (1986)
Mahrhold, S., Rummel, A., Bigalke, H., Davletov, B. & Binz, T. The synaptic vesicle protein 2C mediates the uptake of botulinum neurotoxin A into phrenic nerves. FEBS Lett. 580, 2011–2014 (2006)
Dong, M. et al. SV2 is the protein receptor for botulinum neurotoxin A. Science 312, 592–596 (2006)
Nishiki, T. et al. The high-affinity binding of Clostridium botulinum type B neurotoxin to synaptotagmin II associated with gangliosides GT1b/GD1a. FEBS Lett. 378, 253–257 (1996)
Dong, M. et al. Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J. Cell Biol. 162, 1293–1303 (2003)
Rummel, A., Karnath, T., Henke, T., Bigalke, H. & Binz, T. Synaptotagmins I and II act as nerve cell receptors for botulinum neurotoxin G. J. Biol. Chem. 279, 30865–30870 (2004)
Nishiki, T. et al. Binding of botulinum type B neurotoxin to Chinese hamster ovary cells transfected with rat synaptotagmin II cDNA. Neurosci. Lett. 208, 105–108 (1996)
Swaminathan, S. & Eswaramoorthy, S. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nature Struct. Biol. 7, 693–699 (2000)
Breidenbach, M. A. & Brunger, A. T. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature 432, 925–929 (2004)
Simpson, L. L. Identification of the major steps in botulinum toxin action. Annu. Rev. Pharmacol. Toxicol. 44, 167–193 (2004)
Jayaraman, S., Eswaramoorthy, S., Kumaran, D. & Swaminathan, S. Common binding site for disialyllactose and tri-peptide in C-fragment of tetanus neurotoxin. Proteins 61, 288–295 (2005)
Lacy, D. B., Tepp, W., Cohen, A. C., DasGupta, B. R. & Stevens, R. C. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nature Struct. Biol. 5, 898–902 (1998)
Schiavo, G., Matteoli, M. & Montecucco, C. Neurotoxins affecting neuroexocytosis. Physiol. Rev. 80, 717–766 (2000)
Kitamura, M., Takamiya, K., Aizawa, S. & Furukawa, K. Gangliosides are the binding substances in neural cells for tetanus and botulinum toxins in mice. Biochim. Biophys. Acta 1441, 1–3 (1999)
Kozaki, S., Kamata, Y., Watarai, S., Nishiki, T. & Mochida, S. Ganglioside GT1b as a complementary receptor component for Clostridium botulinum neurotoxins. Microbiol. Pathol. 25, 91–99 (1998)
Rummel, A., Mahrhold, S., Bigalke, H. & Binz, T. The H-CC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Mol. Microbiol. 51, 631–643 (2004)
Ginalski, K., Venclovas, C., Lesyng, B. & Fidelis, K. Structure-based sequence alignment for the beta-trefoil subdomain of the clostridial neurotoxin family provides residue level information about the putative ganglioside binding site. FEBS Lett. 482, 119–124 (2000)
Yowler, B. C., Kensinger, R. D. & Schengrund, C. L. Botulinum neurotoxin A activity is dependent upon the presence of specific gangliosides in neuroblastoma cells expressing synaptotagmin I. J. Biol. Chem. 277, 32815–32819 (2002)
Rummel, A., Bade, S., Alves, J., Bigalke, H. & Binz, T. Two carbohydrate binding sites in the H-cc-domain of tetanus neurotoxin are required for toxicity. J. Mol. Biol. 326, 835–847 (2003)
Fotinou, C. et al. The crystal structure of tetanus toxin Hc fragment complexed with a synthetic GT1b analogue suggests cross-linking between ganglioside receptors and the toxin. J. Biol. Chem. 276, 32274–32281 (2001)
Weis, W. I. Cell-surface carbohydrate recognition by animal and viral lectins. Curr. Opin. Struct. Biol. 7, 624–630 (1997)
Eswaramoorthy, S., Kumaran, D., Keller, J. & Swaminathan, S. Role of metals in the biological activity of Clostridium botulinum neurotoxins. Biochemistry 43, 2209–2216 (2004)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 276, 307–326 (1997)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 (1997)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Cheng, Y. et al. Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain. Protein Sci. 13, 2665–2672 (2004)
Sutton, R. B., Ernst, J. A. & Brunger, A. T. Crystal structure of the cytosolic C2A–C2B domains of synaptotagmin III. Implications for Ca2+-independent SNARE complex interaction. J. Cell Biol. 147, 589–598 (1999)
Acknowledgements
We thank E. Abola, K. Saikatendu and J. Ng for discussions and A. Walker for assistance with manuscript preparation. This work was supported by a grant from the Pacific Southwest Regional Center of Excellence (R.C.S. and E.A.J.), and grants from the NIH/NIAID (to E.R.C.). E.R.C. and E.A.J. acknowledge membership of and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases. E.R.C. is an Investigator of the Howard Hughes Medical Institute. Portions of this research were carried out at the SSRL, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Author Contributions J.W.A., Q.C. (crystallography) and M.D. (molecular biology) contributed equally to this work and co-wrote the paper. All authors discussed the results and commented upon the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Atomic coordinates and experimental structure factors of the BoNT/B–Syt II ectodomain complex have been deposited in the RCSB Protein Data Bank (PDB) under accession code 2NP0. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Table 1 and 2 and Supplementary Figures 1-7 with legends.
Rights and permissions
About this article
Cite this article
Chai, Q., Arndt, J., Dong, M. et al. Structural basis of cell surface receptor recognition by botulinum neurotoxin B . Nature 444, 1096–1100 (2006). https://doi.org/10.1038/nature05411
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05411
This article is cited by
-
Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2
Nature Communications (2023)
-
Split luciferase-based assay to detect botulinum neurotoxins using hiPSC-derived motor neurons
Communications Biology (2023)
-
Molecular landscape of BoNT/B bound to a membrane-inserted synaptotagmin/ganglioside complex
Cellular and Molecular Life Sciences (2022)
-
Bioinformatic discovery of a toxin family in Chryseobacterium piperi with sequence similarity to botulinum neurotoxins
Scientific Reports (2019)
-
Affinity biosensors using recombinant native membrane proteins displayed on exosomes: application to botulinum neurotoxin B receptor
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.