Abstract
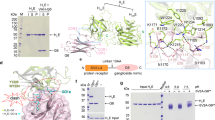
Botulinum neurotoxins (BoNTs) are produced by Clostridium botulinum and cause the neuroparalytic syndrome of botulism. With a lethal dose of 1 ng kg-1, they pose a biological hazard to humans and a serious potential bioweapon threat1. BoNTs bind with high specificity at neuromuscular junctions and they impair exocytosis of synaptic vesicles containing acetylcholine through specific proteolysis of SNAREs (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptors), which constitute part of the synaptic vesicle fusion machinery2,3. The molecular details of the toxin–cell recognition have been elusive. Here we report the structure of a BoNT in complex with its protein receptor: the receptor-binding domain of botulinum neurotoxin serotype B (BoNT/B) bound to the luminal domain of synaptotagmin II, determined at 2.15 Å resolution. On binding, a helix is induced in the luminal domain which binds to a saddle-shaped crevice on a distal tip of BoNT/B. This crevice is adjacent to the non-overlapping ganglioside-binding site of BoNT/B. Synaptotagmin II interacts with BoNT/B with nanomolar affinity, at both neutral and acidic endosomal pH. Biochemical and neuronal ex vivo studies of structure-based mutations indicate high specificity and affinity of the interaction, and high selectivity of BoNT/B among synaptotagmin I and II isoforms. Synergistic binding of both synaptotagmin and ganglioside imposes geometric restrictions on the initiation of BoNT/B translocation after endocytosis. Our results provide the basis for the rational development of preventive vaccines or inhibitors against these neurotoxins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Arnon, S. S. et al. Botulinum toxin as a biological weapon: medical and public health management. J. Am. Med. Assoc. 285, 1059–1070 (2001)
Schiavo, G. et al. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature 359, 832–835 (1992)
Chen, Y. A., Scales, S. J., Patel, S. M., Doung, Y. C. & Scheller, R. H. SNARE complex formation is triggered by Ca2+ and drives membrane fusion. Cell 97, 165–174 (1999)
Montecucco, C. & Schiavo, G. Structure and function of tetanus and botulinum neurotoxins. Q. Rev. Biophys. 28, 423–472 (1995)
Montecucco, C., Rossetto, O. & Schiavo, G. Presynaptic receptor arrays for clostridial neurotoxins. Trends Microbiol. 12, 442–446 (2004)
Rummel, A., Karnath, T., Henke, T., Bigalke, H. & Binz, T. Synaptotagmins I and II act as nerve cell receptors for botulinum neurotoxin G. J. Biol. Chem. 279, 30865–30870 (2004)
Nishiki, T. et al. The high-affinity binding of Clostridium botulinum type B neurotoxin to synaptotagmin II associated with gangliosides GT1b/GD1a. FEBS Lett. 378, 253–257 (1996)
Dong, M. et al. SV2 is the protein receptor for botulinum neurotoxin A. Science 312, 592–596 (2006)
Dong, M. et al. Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J. Cell Biol. 162, 1293–1303 (2003)
Mahrhold, S., Rummel, A., Bigalke, H., Davletov, B. & Binz, T. The synaptic vesicle protein 2C mediates the uptake of botulinum neurotoxin A into phrenic nerves. FEBS Lett. 580, 2011–2014 (2006)
Pang, Z. P. et al. Synaptotagmin-2 is essential for survival and contributes to Ca+2-triggering of neurotransmitter release in central and neuromuscular synapses. J. Neurosci. (in the press).
Eswaramoorthy, S., Kumaran, D. & Swaminathan, S. Crystallographic evidence for doxorubicin binding to the receptor-binding site in Clostridium botulinum neurotoxin B. Acta Crystallogr. D Biol. Crystallogr. 57, 1743–1746 (2001)
Swaminathan, S. & Eswaramoorthy, S. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nature Struct. Biol. 7, 693–699 (2000)
Rummel, A., Mahrhold, S., Bigalke, H. & Binz, T. The HCC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Mol. Microbiol. 51, 631–643 (2004)
Jayaraman, S., Eswaramoorthy, S., Ahmed, S. A., Smith, L. A. & Swaminathan, S. N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B. Biochem. Biophys. Res. Commun. 330, 97–103 (2005)
Perozzo, R., Folkers, G. & Scapozza, L. Thermodynamics of protein–ligand interactions: history, presence, and future aspects. J. Recept. Signal Transduct. Res. 24, 1–52 (2004)
Stites, W. E. Protein–protein interactions: interface structure, binding thermodynamics, and mutational analysis. Chem. Rev. 97, 1233–1250 (1997)
Habermann, E., Dreyer, F. & Bigalke, H. Tetanus toxin blocks the neuromuscular transmission in vitro like botulinum A toxin. Naunyn Schmiedebergs Arch. Pharmacol. 311, 33–40 (1980)
Südhof, T. C. Synaptotagmins: why so many? J. Biol. Chem. 277, 7629–7632 (2002)
Rummel, A. et al. Identification of the protein receptor binding site of botulinum neurotoxins B and G proves the double receptor concept. Proc. Natl Acad. Sci. USA (in the press).
Montecucco, C. How do tetanus and botulinum neurotoxins bind to neuronal membranes? Trends Biochem. Sci. 11, 314–317 (1986)
Kozaki, S., Kamata, Y., Watarai, S., Nishiki, T. & Mochida, S. Ganglioside GT1b as a complementary receptor component for Clostridium botulinum neurotoxins. Microb. Pathog. 25, 91–99 (1998)
Rummel, A., Bade, S., Alves, J., Bigalke, H. & Binz, T. Two carbohydrate binding sites in the HCC-domain of tetanus neurotoxin are required for toxicity. J. Mol. Biol. 326, 835–847 (2003)
Koriazova, L. K. & Montal, M. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nature Struct. Biol. 10, 13–18 (2003)
Hoch, D. H. et al. Channels formed by botulinum, tetanus, and diphtheria toxins in planar lipid bilayers: relevance to translocation of proteins across membranes. Proc. Natl Acad. Sci. USA 82, 1692–1696 (1985)
Dolinsky, T. J., Nielsen, J. E., McCammon, J. A. & Baker, N. A. PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004)
Li, H., Robertson, A. D. & Jensen, J. H. Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721 (2005)
McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61, 458–464 (2005)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Acknowledgements
We thank P. Adams and J. Zhou for critical reading of the manuscript, and the staff of beamlines 9-1 at the Stanford Synchrotron Radiation Laboratory (SSRL) and beam line 8.2.2 at the Advanced Light Source (ALS) for help during data collection; H. Bigalke for providing the mouse phrenic nerve facility; and T. Henke and C. Knorr for technical assistance. The SSRL is a national user facility operated by Stanford University on behalf of the US Department of Energy (Office of Basic Energy Sciences). The SSRL Structural Molecular Biology Program is supported by the Department of Energy (Office of Biological and Environmental Research), and by the National Institutes of Health (National Center for Research Resources, Biomedical Technology Program), and the National Institute of General Medical Sciences. The ALS is supported by the Office of Energy Research (Office of Basic Energy Sciences, Material Sciences Division) of the US Department of Energy at Lawrence Berkeley National Laboratory. Support by the Department of Defense and Defense Threat Reduction Agency (to A.T.B.) and by a Deutsche Forschungsgemeinschaft grant (to T.B.) is acknowledged. The atomic coordinates and structure factors of the HCB–Syt-II complex are deposited in the Protein Data Bank under accession code 2NM1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The atomic coordinates and structure factors of the HCB–Syt-II complex are deposited in the Protein Data Bank under accession code 2NM1. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Materials and Methods, Supplementary Tables 1-2 and Supplementary Figures 1-5 with legends
Rights and permissions
About this article
Cite this article
Jin, R., Rummel, A., Binz, T. et al. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature 444, 1092–1095 (2006). https://doi.org/10.1038/nature05387
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05387
This article is cited by
-
Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A
Nature Communications (2024)
-
Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2
Nature Communications (2023)
-
Split luciferase-based assay to detect botulinum neurotoxins using hiPSC-derived motor neurons
Communications Biology (2023)
-
Structure of the glucosyltransferase domain of TcdA in complex with RhoA provides insights into substrate recognition
Scientific Reports (2022)
-
Molecular landscape of BoNT/B bound to a membrane-inserted synaptotagmin/ganglioside complex
Cellular and Molecular Life Sciences (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.