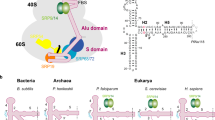
Abstract
The prokaryotic signal recognition particle (SRP) targets membrane proteins into the inner membrane1,2,3,4. It binds translating ribosomes and screens the emerging nascent chain for a hydrophobic signal sequence, such as the transmembrane helix of inner membrane proteins. If such a sequence emerges, the SRP binds tightly, allowing the SRP receptor to lock on. This assembly delivers the ribosome-nascent chain complex to the protein translocation machinery in the membrane. Using cryo-electron microscopy and single-particle reconstruction, we obtained a 16 Å structure of the Escherichia coli SRP in complex with a translating E. coli ribosome containing a nascent chain with a transmembrane helix anchor. We also obtained structural information on the SRP bound to an empty E. coli ribosome. The latter might share characteristics with a scanning SRP complex, whereas the former represents the next step: the targeting complex ready for receptor binding. High-resolution structures of the bacterial ribosome and of the bacterial SRP components are available, and their fitting explains our electron microscopic density. The structures reveal the regions that are involved in complex formation, provide insight into the conformation of the SRP on the ribosome and indicate the conformational changes that accompany high-affinity SRP binding to ribosome nascent chain complexes upon recognition of the signal sequence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Keenan, R. J., Freymann, D. M., Stroud, R. M. & Walter, P. The signal recognition particle. Annu. Rev. Biochem. 70, 755–775 (2001)
Nagai, K. et al. Structure, function and evolution of the signal recognition particle. EMBO J. 22, 3479–3485 (2003)
Doudna, J. A. & Batey, R. T. Structural insights into the signal recognition particle. Annu. Rev. Biochem. 73, 539–557 (2004)
Ulbrandt, N. D., Newitt, J. A. & Bernstein, H. D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88, 187–196 (1997)
Walter, P. & Blobel, G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 299, 691–698 (1982)
Gu, S. Q., Peske, F., Wieden, H. J., Rodnina, M. V. & Wintermeyer, W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA 9, 566–573 (2003)
Pool, M. R., Stumm, J., Fulga, T. A., Sinning, I. & Dobberstein, B. Distinct modes of signal recognition particle interaction with the ribosome. Science 297, 1345–1348 (2002)
Keenan, R. J., Freymann, D. M., Walter, P. & Stroud, R. M. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94, 181–191 (1998)
Halic, M. et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814 (2004)
Mitra, K. et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324 (2005)
Valent, Q. A. et al. Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol. Microbiol. 25, 53–64 (1997)
Buskiewicz, I. et al. Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc. Natl Acad. Sci. USA 101, 7902–7906 (2004)
Raine, A., Ivanova, N., Wikberg, J. E. & Ehrenberg, M. Simultaneous binding of trigger factor and signal recognition particle to the E. coli ribosome. Biochimie 86, 495–500 (2004)
Schuwirth, B. S. et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834 (2005)
Wriggers, W., Milligan, R. A. & McCammon, J. A. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125, 185–195 (1999)
Padmanabhan, S. & Freymann, D. M. The conformation of bound GMPPNP suggests a mechanism for gating the active site of the SRP GTPase. Structure 9, 859–867 (2001)
Rosendal, K. R., Wild, K., Montoya, G. & Sinning, I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc. Natl Acad. Sci. USA 100, 14701–14706 (2003)
Rosenblad, M. A., Gorodkin, J., Knudsen, B., Zwieb, C. & Samuelsson, T. SRPDB: Signal Recognition Particle Database. Nucleic Acids Res. 31, 363–364 (2003)
Lindahl, E., Azuara, C., Koehl, P. & Delarue, M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 34, W52–W56 (2006)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54, 905–921 (1998)
Wild, K., Halic, M., Sinning, I. & Beckmann, R. SRP meets the ribosome. Nature Struct. Mol. Biol. 11, 1049–1053 (2004)
Ullers, R. S. et al. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell Biol. 161, 679–684 (2003)
Eisner, G., Moser, M., Schäfer, U., Beck, K. & Müller, M. Alternate recruitment of signal recognition particle and trigger factor to the signal sequence of a growing nascent polypeptide. J. Biol. Chem. 281, 7172–7179 (2006)
Ferbitz, L. et al. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431, 590–596 (2004)
Rinke-Appel, J. et al. Crosslinking of 4.5S RNA to the Escherichia coli ribosome in the presence or absence of the protein Ffh. RNA 8, 612–625 (2002)
Conway, J. F. & Steven, A. C. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 128, 106–118 (1999)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Valle, M. et al. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 21, 3557–3567 (2002)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Bacher, G., Lütcke, H., Jungnickel, B., Rapoport, T. A. & Dobberstein, B. Regulation by the ribosome of the GTPase of the signal-recognition particle during protein targeting. Nature 381, 248–251 (1996)
Acknowledgements
The authors would like to thank all members of the Ban laboratory for discussions and help with programs, and C. Frick, L. Verschragen (deceased) and J. Onderwater for technical assistance. W. Wintermeyer provided pET24a_Ffh. T. Shaikh and J. Frank are acknowledged for a script for supervised classification. We thank the Electron Microscopy Center Zürich (EMEZ) for support. C.S. was supported by a postdoctoral fellowship from the Ernst Schering Research Foundation. R.I.K. was supported by a VENI grant from the Netherlands Organisation for Scientific Research (NWO). This work was supported by the Swiss National Science Foundation (SNSF), the NCCR Structural Biology program of the SNSF, and a Young Investigator grant from the Human Frontier Science Program (to N.B.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Coordinates of the atomic model of SRP have been deposited in the Protein Data Bank under the accession code 2iy3. The cryo-EM maps have been deposited in the 3D-EM database (EMBL-European Bioinformatics Institute, Cambridge, UK) under accession numbers EMD-1250 and EMD-1251. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains the Supplementary Results and Supplementary Discussion, Supplementary Methods, Supplementary Figures and Legends 1–4, Supplementary Table 1 and additional references. (PDF 338 kb)
Rights and permissions
About this article
Cite this article
Schaffitzel, C., Oswald, M., Berger, I. et al. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature 444, 503–506 (2006). https://doi.org/10.1038/nature05182
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05182
This article is cited by
-
The Principles of Protein Targeting and Transport Across Cell Membranes
The Protein Journal (2019)
-
The signal recognition particle contacts uL23 and scans substrate translation inside the ribosomal tunnel
Nature Microbiology (2017)
-
Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon
Nature Communications (2016)
-
A structural ensemble of a ribosome–nascent chain complex during cotranslational protein folding
Nature Structural & Molecular Biology (2016)
-
Signal-sequence induced conformational changes in the signal recognition particle
Nature Communications (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.