Abstract
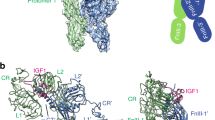
The insulin receptor is a phylogenetically ancient tyrosine kinase receptor found in organisms as primitive as cnidarians and insects. In higher organisms it is essential for glucose homeostasis1, whereas the closely related insulin-like growth factor receptor (IGF-1R) is involved in normal growth and development2. The insulin receptor is expressed in two isoforms, IR-A and IR-B; the former also functions as a high-affinity receptor for IGF-II and is implicated, along with IGF-1R, in malignant transformation3. Here we present the crystal structure at 3.8 Å resolution of the IR-A ectodomain dimer, complexed with four Fabs from the monoclonal antibodies 83-7 and 83-14 (ref. 4), grown in the presence of a fragment of an insulin mimetic peptide5. The structure reveals the domain arrangement in the disulphide-linked ectodomain dimer, showing that the insulin receptor adopts a folded-over conformation that places the ligand-binding regions in juxtaposition. This arrangement is very different from previous models6. It shows that the two L1 domains are on opposite sides of the dimer, too far apart to allow insulin to bind both L1 domains simultaneously as previously proposed7. Instead, the structure implicates the carboxy-terminal surface of the first fibronectin type III domain as the second binding site involved in high-affinity binding.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kitamura, T., Kahn, C. R. & Accili, D. Insulin receptor knockout mice. Annu. Rev. Physiol. 65, 313–332 (2003)
Adams, T. E., Epa, V. C., Garrett, T. P. J. & Ward, C. W. Structure and function of the type 1 insulin-like growth factor receptor. Cell. Mol. Life Sci. 57, 1050–1093 (2000)
Denley, A., Wallace, J. C., Cosgrove, L. J. & Forbes, B. E. The insulin receptor isoform exon 11- (IR-A) in cancer and other diseases: a review. Horm. Metab. Res. 35, 778–785 (2003)
Soos, M. A. et al. Monoclonal antibodies reacting with multiple epitopes on the human insulin receptor. Biochem. J. 235, 199–208 (1986)
Schaffer, L. et al. Assembly of high-affinity insulin receptor agonists and antagonists from peptide building blocks. Proc. Natl Acad. Sci. USA 100, 4435–4439 (2003)
De Meyts, P. & Whittaker, J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nature Rev. Drug Discov. 1, 769–783 (2002)
Ottensmeyer, F. P., Beniac, D. R., Luo, R. Z. T. & Yip, C. C. Mechanism of transmembrane signaling: insulin binding and the insulin receptor. Biochemistry 39, 12103–12112 (2000). Corrigendum. Ibid. 40, 6988
Sparrow, L. G. et al. The disulfide bonds in the C-terminal domains of the human insulin receptor ectodomain. J. Biol. Chem. 272, 29460–29467 (1997)
Peng, K. et al. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 3, 35–60 (2005)
Lou, M. et al. Crystal structure of the first three domains of the human insulin receptor reveals major differences from the IGF-1 receptor in the regions governing ligand specificity. Proc. Natl Acad. Sci. USA 103, 12429–12434 (2006)
Garrett, T. P. J. et al. Crystal structure of the first three domains of the type-1 insulin-like growth factor receptor. Nature 394, 395–399 (1998)
Kadowaki, H. et al. Mutagenesis of lysine 460 in the human insulin receptor: effects upon receptor recycling and cooperative interactions among binding sites. J. Biol. Chem. 265, 21285–21296 (1990)
Marino-Buslje, C., Martin-Martinez, M., Mizuguchi, K., Siddle, K. & Blundell, T. L. The insulin receptor: from protein sequence to structure. Biochem. Soc. Trans. 27, 715–726 (1999)
Schaefer, E. M., Erickson, H. P., Federwisch, M., Wollmer, A. & Ellis, L. Structural organization of the human insulin receptor ectodomain. J. Biol. Chem. 267, 23393–23402 (1992)
Cheatham, B. & Kahn, C. R. Cysteine 647 in the insulin receptor is required for normal covalent interaction between α- and β-subunits and signal transduction. J. Biol. Chem. 267, 7108–7115 (1992)
Tulloch, P. A. et al. Single-molecule imaging of human insulin receptor ectodomain and its Fab complexes. J. Struct. Biol. 125, 11–18 (1999)
Burgess, A. W. et al. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell 12, 541–552 (2003)
Lee, J., Pilch, P. F., Shoelson, S. E. & Scarlata, S. F. Conformational changes of the insulin receptor upon insulin binding and activation as monitored by fluorescence spectroscopy. Biochemistry 36, 2701–2708 (1997)
Florke, R. R. et al. Hormone-triggered conformational changes within the insulin-receptor ectodomain: requirement for transmembrane anchors. Biochem. J. 360, 189–198 (2001)
Yip, C. C. & Ottensmeyer, P. Three-dimensional structural interactions of insulin and its receptor. J. Biochem. 278, 27329–27332 (2003)
Surinya, K. H. et al. Role of insulin receptor dimerization domains in ligand binding, cooperativity, and modulation by anti-receptor antibodies. J. Biol. Chem. 277, 16718–16725 (2002)
Kurose, T. et al. Cross-linking of a B25 azidophenylalanine insulin derivative to the carboxy-terminal region of the α-subunit of the insulin receptor. Identification of a new insulin-binding domain in the insulin receptor. J. Biol. Chem. 269, 29190–29197 (1994)
Kristensen, C., Wiberg, F. C. & Andersen, A. S. Specificity of insulin and insulin-like growth factor I receptors investigated using chimeric mini-receptors—role of C-terminal of receptor α subunit. J. Biol. Chem. 274, 37351–37356 (1999)
Schaffer, L. A model for insulin binding to the insulin receptor. Eur. J. Biochem. 221, 1127–1132 (1994)
De Meyts, P. The structural basis of insulin and IGF-1 receptor binding and negative co-operativity, and its relevance to mitogenic versus metabolic signaling. Diabetologia 37 (suppl. 2), S135–S148 (1994)
De Meyts, P. Insulin and its receptor: structure, function and evolution. BioEssays 26, 1351–1362 (2004)
Zhang, B. & Roth, R. A. A region of the insulin receptor important for ligand binding (residues 450–601) is recognized by patient's autoimmune antibodies and inhibitory monoclonal antibodies. Proc. Natl Acad. Sci. USA 88, 9858–9862 (1991)
Schumacher, R. et al. Signaling-competent receptor chimeras allow mapping of major insulin receptor binding domain determinants. J. Biol. Chem. 268, 1087–1094 (1993)
Fabry, M. et al. Detection of a new hormone contact site within the insulin receptor ectodomain by the use of a novel photoreactive insulin. J. Biol. Chem. 267, 8950–8956 (1992)
Hoyne, P. A. et al. High affinity insulin binding by soluble insulin receptor extracellular domain fused to a leucine zipper. FEBS Lett. 479, 15–18 (2000)
Acknowledgements
We thank K. Siddle for the 83-7 and 83-14 monoclonal antibody cell lines; O. Dolezal for advice on cloning and sequencing the monoclonal antibody variable region cDNAs; B. Van Donkelaar for technical crystallography inputs; and L. Lu, L. Cheong and T. Phan for contributions to protein production. We acknowledge the help provided by beamline staff at both the Advanced Photon Source and the Photon Factory. This work was supported by the Australian Synchrotron Research Program, which is funded by the Commonwealth of Australia under the Major National Research Facilities Program. Additional financial support was provided under the Generic Technology component of the Industry Research and Development Act 1986, from Biota Diabetes Research Pty Ltd.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Atomic coordinates of the IR-Fab complex have been deposited in the Protein Data Bank with accession number 2DTG. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
Models of insulin/IR binding. This file contains an overview of the models for insulin binding to the insulin receptor plus references. (DOC 31 kb)
Supplementary Methods
This file contains expanded methods details and references. (DOC 43 kb)
Supplementary Table 1
Diffraction data and refinement statistics. (DOC 35 kb)
Supplementary Figure 1
Schematic diagram of the insulin receptor dimer. (JPG 195 kb)
Supplementary Figure 2
Sequence alignment of the ectodomains of human insulin receptor (IR, exon 11- isoform) and human IGF1R (IGF-1R). (JPG 531 kb)
Supplementary Figure 3
Polypeptide fold for the IR monomer showing the relative arrangement of domains. (JPG 270 kb)
Supplementary Figure 4
Segment of difference electron density lying across the face of domain L1. (JPG 302 kb)
Supplementary Figure 5
Nucleotide and derived amino acid sequences of the light and heavy chain variable regions of the Fab from the anti-insulin receptor monoclonal antibody 83-7. (DOC 28 kb)
Supplementary Figure 6
Nucleotide and derived amino acid sequences of the light and heavy chain variable regions of the Fab from the anti-insulin receptor monoclonal antibody 83-14. (DOC 38 kb)
Supplementary Figure 7
McKern et al. Supplementary Figure S7. Cartoon of insulin binding to membrane-anchored IR. (JPG 4885 kb)
Supplementary Figure 8
Stereo images of 2Fo-Fc maps contoured at 1 σ contour level produced by BUSTER-TNT. (DOC 1833 kb)
Rights and permissions
About this article
Cite this article
McKern, N., Lawrence, M., Streltsov, V. et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature 443, 218–221 (2006). https://doi.org/10.1038/nature05106
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05106
This article is cited by
-
A stepwise activation model for the insulin receptor
Experimental & Molecular Medicine (2023)
-
Recombinant Insulin-Like Growth Factor 1 Dimers: Receptor Binding Affinities and Activation Abilities
International Journal of Peptide Research and Therapeutics (2023)
-
Synergistic activation of the insulin receptor via two distinct sites
Nature Structural & Molecular Biology (2022)
-
An aptamer agonist of the insulin receptor acts as a positive or negative allosteric modulator, depending on its concentration
Experimental & Molecular Medicine (2022)
-
Activation of the human insulin receptor by non-insulin-related peptides
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.