Abstract
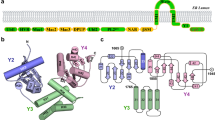
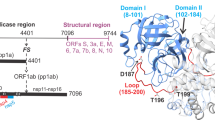
Hepatitis C virus is a major global health problem affecting an estimated 170 million people worldwide1. Chronic infection is common and can lead to cirrhosis and liver cancer. There is no vaccine available and current therapies have met with limited success2. The viral RNA genome encodes a polyprotein that includes two proteases essential for virus replication3,4. The NS2-3 protease mediates a single cleavage at the NS2/NS3 junction, whereas the NS3-4A protease cleaves at four downstream sites in the polyprotein. NS3-4A is characterized as a serine protease with a chymotrypsin-like fold5,6, but the enzymatic mechanism of the NS2-3 protease remains unresolved7,8,9. Here we report the crystal structure of the catalytic domain of the NS2-3 protease at 2.3 Å resolution. The structure reveals a dimeric cysteine protease with two composite active sites. For each active site, the catalytic histidine and glutamate residues are contributed by one monomer, and the nucleophilic cysteine by the other. The carboxy-terminal residues remain coordinated in the two active sites, predicting an inactive post-cleavage form. Proteolysis through formation of a composite active site occurs in the context of the viral polyprotein expressed in mammalian cells. These features offer unexpected insights into polyprotein processing by hepatitis C virus and new opportunities for antiviral drug design.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Alter, H. J. & Seeff, L. B. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20, 17–35 (2000)
Hoofnagle, J. H. Course and outcome of hepatitis C. Hepatology 36, S21–S29 (2002)
Bartenschlager, R., Frese, M. & Pietschmann, T. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63, 71–180 (2004)
Lindenbach, B. D. & Rice, C. M. in Fields Virology (eds Knipe, D. M. et al.) 991–1041 (Lippincott Williams & Wilkins, Philadelphia, Pennsylvania, 2001)
Love, R. A. et al. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87, 331–342 (1996)
Kim, J. L. et al. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87, 343–355 (1996)
Grakoui, A., McCourt, D. W., Wychowski, C., Feinstone, S. M. & Rice, C. M. A second hepatitis C virus-encoded proteinase. Proc. Natl Acad. Sci. USA 90, 10583–10587 (1993)
Hijikata, M. et al. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67, 4665–4675 (1993)
Wu, Z., Yao, N., Le, H. V. & Weber, P. C. Mechanism of autoproteolysis at the NS2–NS3 junction of the hepatitis C virus polyprotein. Trends Biochem. Sci. 23, 92–94 (1998)
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993)
Pickersgill, R. W., Harris, G. W. & Garman, E. Structure of monoclinic papain at 1.60 Å resolution. Acta Crystallogr. B 48, 49–66 (1992)
Mosimann, S. C., Cherney, M. M., Sia, S., Plotch, S. & James, M. N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 273, 1032–1047 (1997)
Tong, L., Wengler, G. & Rossmann, M. G. Refined structure of Sindbis virus core protein and comparison with other chymotrypsin-like serine proteinase structures. J. Mol. Biol. 230, 228–247 (1993)
McPhalen, C. A. & James, M. N. Structural comparison of two serine proteinase-protein inhibitor complexes: eglin-c-subtilisin Carlsberg and CI-2-subtilisin Novo. Biochemistry 27, 6582–6598 (1988)
Malcolm, B. A. The picornaviral 3C proteinases: cysteine nucleophiles in serine proteinase folds. Protein Sci. 4, 1439–1445 (1995)
Wlodawer, A. & Gustchina, A. Structural and biochemical studies of retroviral proteases. Biochim. Biophys. Acta 1477, 16–34 (2000)
Boatright, K. M. & Salvesen, G. S. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 15, 725–731 (2003)
Thibeault, D., Maurice, R., Pilote, L., Lamarre, D. & Pause, A. In vitro characterization of a purified NS2/3 protease variant of hepatitis C virus. J. Biol. Chem. 276, 46678–46684 (2001)
Pallaoro, M. et al. Characterization of the hepatitis C virus NS2/3 processing reaction by using a purified precursor protein. J. Virol. 75, 9939–9946 (2001)
Urbani, A. et al. The metal binding site of the hepatitis C virus NS3 protease. A spectroscopic investigation. J. Biol. Chem. 273, 18760–18769 (1998)
Gale, M. Jr & Foy, E. M. Evasion of intracellular host defence by hepatitis C virus. Nature 436, 939–945 (2005)
Pietschmann, T. et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl Acad. Sci. USA 103, 7408–7413 (2006)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Weeks, C. M. & Miller, R. The design and implementation of SnB v2.0. J. Appl. Crystallogr. 32, 120–124 (1999)
Collaborative Computational Project No. 4, The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Santolini, E., Pacini, L., Fipaldini, C., Migliaccio, G. & Monica, N. The NS2 protein of hepatitis C virus is a transmembrane polypeptide. J. Virol. 69, 7461–7471 (1995)
Yamaga, A. K. & Ou, J. H. Membrane topology of the hepatitis C virus NS2 protein. J. Biol. Chem. 277, 33228–33234 (2002)
Acknowledgements
We would like to thank R. MacKinnon, S. Darst and H. Mueller for the use of X-ray diffractometers, related equipment and software. We appreciate access to beamlines X9A and X29 of the National Synchrotron Light Source (NSLS) at the Brookhaven National Laboratory and acknowledge the assistance of the NSLS staff. We wish to thank T. Tellinghuisen for assistance with data collection. D. Jeruzalmi provided the program msf_similarity_to_pdb and LOOSENGRASP. We would like to thank S. Burley, S. Darst, L. Dustin, M. Evans, C. Jones, C. Murray, B. Lindenbach, G. Randall and T. Tellinghuisen for input on the manuscript, and K. Kuo and S. You for help with artwork. I.C.L. was supported by fellowships from the Swiss National Science Foundation, the Roche Research Foundation and the Swiss Society for Medical-Biological Grants. J.M. was supported as a Merck Fellow of the Life Sciences Research Foundation. Additional financial support for this work came from grants from the National Institutes of Health and the Greenberg Medical Research Institute (C.M.R.). Author Contributions I.C.L., J.M. and C.M.R. conceived the experiments. I.C.L. generated all reagents, materials, proteins and crystals with assistance from J.M. All data collection and processing was done by J.M. and I.C.L. Model building and refinement were performed by I.C.L. and J.M. Cross-linking and mammalian expression studies were carried out by I.C.L. and T.G.D. The manuscript was written by I.C.L., J.M. and C.M.R.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The atomic coordinates for this structure have been deposited in the Protein Data Bank under accession code 2HD0. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Summary of crystal parameters, data collection and refinement statistics. (PDF 86 kb)
Supplementary Figure 1
Asymmetric unit of the native NS2pro crystal. (PDF 534 kb)
Supplementary Figure 2
Electron density maps of the linker polypeptide, the active site, the cis-proline residue, and a detergent molecule. (PDF 705 kb)
Supplementary Figure 3
Sequence conservation at the solvent-accessible surface of NS2pro. (PDF 235 kb)
Supplementary Figure 4
Dimerization of purified NS2pro in solution with a chemical crosslinker. (PDF 101 kb)
Supplementary Figure Legends
This file contains legends to the Supplementary Figures. (DOC 26 kb)
Supplementary Data
Supplementary Data nature04975-s7.doc This file contains a detailed description of the methods, and additional references. (DOC 68 kb)
Rights and permissions
About this article
Cite this article
Lorenz, I., Marcotrigiano, J., Dentzer, T. et al. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442, 831–835 (2006). https://doi.org/10.1038/nature04975
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04975
This article is cited by
-
Viral genome wide association study identifies novel hepatitis C virus polymorphisms associated with sofosbuvir treatment failure
Nature Communications (2021)
-
Hepatitis C Virus NS2 Protein Suppresses RNA Interference in Cells
Virologica Sinica (2020)
-
Phosphatidylserine-Specific Phospholipase A1 is the Critical Bridge for Hepatitis C Virus Assembly
Virologica Sinica (2019)
-
Pharmacophore anchor models of flaviviral NS3 proteases lead to drug repurposing for DENV infection
BMC Bioinformatics (2017)
-
Structure of the NS2B-NS3 protease from Zika virus after self-cleavage
Nature Communications (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.