Abstract
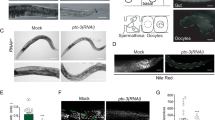
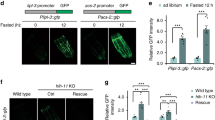
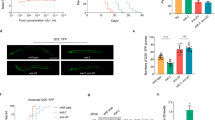
The sterol regulatory element binding protein (SREBP) family of transcription activators are critical regulators of cholesterol and fatty acid homeostasis1,2. We previously demonstrated that human SREBPs bind the CREB-binding protein (CBP)/p300 acetyltransferase KIX domain and recruit activator-recruited co-factor (ARC)/Mediator co-activator complexes through unknown mechanisms3,4,5. Here we show that SREBPs use the evolutionarily conserved ARC105 (also called MED15) subunit to activate target genes. Structural analysis of the SREBP-binding domain in ARC105 by NMR revealed a three-helix bundle with marked similarity to the CBP/p300 KIX domain. In contrast to SREBPs, the CREB and c-Myb activators do not bind the ARC105 KIX domain, although they interact with the CBP KIX domain, revealing a surprising specificity among structurally related activator-binding domains. The Caenorhabditis elegans SREBP homologue SBP-1 promotes fatty acid homeostasis by regulating the expression of lipogenic enzymes6,7. We found that, like SBP-1, the C. elegans ARC105 homologue MDT-15 is required for fatty acid homeostasis, and show that both SBP-1 and MDT-15 control transcription of genes governing desaturation of stearic acid to oleic acid. Notably, dietary addition of oleic acid significantly rescued various defects of nematodes targeted with RNA interference against sbp-1 and mdt-15, including impaired intestinal fat storage, infertility, decreased size and slow locomotion, suggesting that regulation of oleic acid levels represents a physiologically critical function of SBP-1 and MDT-15. Taken together, our findings demonstrate that ARC105 is a key effector of SREBP-dependent gene regulation and control of lipid homeostasis in metazoans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Brown, M. S. & Goldstein, J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997)
Eberle, D., Hegarty, B., Bossard, P., Ferre, P. & Foufelle, F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 86, 839–848 (2004)
Näär, A. M. et al. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 12, 3020–3031 (1998)
Näär, A. M. et al. Composite coactivator ARC mediates chromatin-directed transcriptional activation. Nature 398, 828–832 (1999)
Oliner, J. D., Andresen, J. M., Hansen, S. K., Zhou, S. & Tjian, R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 10, 2903–2911 (1996)
Kniazeva, M., Crawford, Q. T., Seiber, M., Wang, C. Y. & Han, M. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2, E257 (2004)
McKay, R. M., McKay, J. P., Avery, L. & Graff, J. M. C. elegans: a model for exploring the genetics of fat storage. Dev. Cell 4, 131–142 (2003)
Jump, D. B. Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr. Opin. Lipidol. 13, 155–164 (2002)
Maxfield, F. R. & Tabas, I. Role of cholesterol and lipid organization in disease. Nature 438, 612–621 (2005)
Moller, D. E. & Kaufman, K. D. Metabolic syndrome: a clinical and molecular perspective. Annu. Rev. Med. 56, 45–62 (2005)
Toth, J. I., Datta, S., Athanikar, J. N., Freedman, L. P. & Osborne, T. F. Selective coactivator interactions in gene activation by SREBP-1a and -1c. Mol. Cell. Biol. 24, 8288–8300 (2004)
Novatchkova, M. & Eisenhaber, F. Linking transcriptional mediators via the GACKIX domain super family. Curr. Biol. 14, R54–R55 (2004)
Kato, Y., Habas, R., Katsuyama, Y., Näär, A. M. & He, X. A component of the ARC/Mediator complex required for TGF-β/Nodal signalling. Nature 418, 641–646 (2002)
Yang, F., DeBeaumont, R., Zhou, S. & Näär, A. M. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc. Natl Acad. Sci. USA 101, 2339–2344 (2004)
Chrivia, J. C. et al. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365, 855–859 (1993)
Dai, P. et al. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 10, 528–540 (1996)
Radhakrishnan, I. et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell 91, 741–752 (1997)
Zor, T., De Guzman, R. N., Dyson, H. J. & Wright, P. E. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J. Mol. Biol. 337, 521–534 (2004)
Parker, D. et al. Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol. Cell 2, 353–359 (1998)
Parker, D. et al. Role of secondary structure in discrimination between constitutive and inducible activators. Mol. Cell. Biol. 19, 5601–5607 (1999)
Goto, N. K., Zor, T., Martinez-Yamout, M., Dyson, H. J. & Wright, P. E. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J. Biol. Chem. 277, 43168–43174 (2002)
De Guzman, R. N., Goto, N. K., Dyson, H. J. & Wright, P. E. Structural basis for cooperative transcription factor binding to the CBP coactivator. J. Mol. Biol. 355, 1005–1013 (2006)
Bourbon, H. M. et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14, 553–557 (2004)
Ashrafi, K. et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272 (2003)
Kamath, R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003)
Tabor, D. E., Kim, J. B., Spiegelman, B. M. & Edwards, P. A. Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-CoA desaturase 1 and 2. J. Biol. Chem. 274, 20603–20610 (1999)
Watts, J. L. & Browse, J. A palmitoyl-CoA-specific delta9 fatty acid desaturase from Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 272, 263–269 (2000)
Watts, J. L. & Browse, J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 99, 5854–5859 (2002)
Taubert, S., Van, M. R., Hansen, M. & Yamamoto, K. R. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 20, 1137–1149 (2006)
Yuan, C. X., Ito, M., Fondell, J. D., Fu, Z. Y. & Roeder, R. G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl Acad. Sci. USA 95, 7939–7944 (1998)
Acknowledgements
We were unable to cite many original papers owing to space constraints. We acknowledge G. Gill, N. Dyson and B. Spiegelman for comments on the manuscript. We thank B. Lüscher, R. Sordella and M. Classon for reagents, N. J. Moerke for his assistance with the fluorescence polarization experiments, and M. Dedmon for providing purified ARC105. A.M.N. is a Dammerman Scholar of the Damon Runyon Cancer Research Foundation. This work was supported by the NIH, the Damon Runyon Cancer Research Foundation, and the Milton Foundation of Harvard University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Atomic coordinates of the amino-terminal region of ARC105 containing the KIX domain has been deposited in the Protein Data Bank with the accession number 2GUT. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Figures 1–9, Supplementary Tables 1 and 2, Supplementary Discussion, Supplementary Methods and additional references. (PDF 572 kb)
Rights and permissions
About this article
Cite this article
Yang, F., Vought, B., Satterlee, J. et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700–704 (2006). https://doi.org/10.1038/nature04942
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04942
This article is cited by
-
Regulation of the RNA polymerase II pre-initiation complex by its associated coactivators
Nature Reviews Genetics (2023)
-
The translational potential of cholesterol-based therapies for neurological disease
Nature Reviews Neurology (2023)
-
A new AMPK isoform mediates glucose-restriction induced longevity non-cell autonomously by promoting membrane fluidity
Nature Communications (2023)
-
Lipid droplets and peroxisomes are co-regulated to drive lifespan extension in response to mono-unsaturated fatty acids
Nature Cell Biology (2023)
-
The CRTC-1 transcriptional domain is required for COMPASS complex-mediated longevity in C. elegans
Nature Aging (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.