Abstract
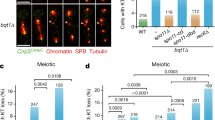
Segregation of homologous maternal and paternal centromeres to opposite poles during meiosis I depends on post-replicative crossing over between homologous non-sister chromatids, which creates chiasmata and therefore bivalent chromosomes. Destruction of sister chromatid cohesion along chromosome arms due to proteolytic cleavage of cohesin's Rec8 subunit by separase resolves chiasmata and thereby triggers the first meiotic division. This produces univalent chromosomes, the chromatids of which are held together by centromeric cohesin that has been protected from separase by shugoshin (Sgo1/MEI-S332) proteins. Here we show in both fission and budding yeast that Sgo1 recruits to centromeres a specific form of protein phosphatase 2A (PP2A). Its inactivation causes loss of centromeric cohesin at anaphase I and random segregation of sister centromeres at the second meiotic division. Artificial recruitment of PP2A to chromosome arms prevents Rec8 phosphorylation and hinders resolution of chiasmata. Our data are consistent with the notion that efficient cleavage of Rec8 requires phosphorylation of cohesin and that this is blocked by PP2A at meiosis I centromeres.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Nasmyth, K. & Haering, C. H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74, 595–648 (2005)
Nasmyth, K. How might cohesin hold sister chromatids together? Phil. Trans. R. Soc. Lond. B 360, 483–496 (2005)
Toth, A. et al. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 103, 1155–1168 (2000)
Rabitsch, K. P. et al. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell 4, 535–548 (2003)
Buonomo, S. B. et al. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103, 387–398 (2000)
Kitajima, T. S., Miyazaki, Y., Yamamoto, M. & Watanabe, Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J. 22, 5643–5653 (2003)
Rabitsch, K. P. et al. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 14, 287–301 (2004)
Kitajima, T. S., Kawashima, S. A. & Watanabe, Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427, 510–517 (2004)
Kerrebrock, A. W., Moore, D. P., Wu, J. S. & Orr-Weaver, T. L. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 83, 247–256 (1995)
Katis, V. L., Galova, M., Rabitsch, K. P., Gregan, J. & Nasmyth, K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 14, 560–572 (2004)
Marston, A. L., Tham, W. H., Shah, H. & Amon, A. A genome-wide screen identifies genes required for centromeric cohesion. Science 303, 1367–1370 (2004)
Bernard, P., Maure, J. F. & Javerzat, J. P. Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nature Cell Biol. 3, 522–526 (2001)
Hauf, S. et al. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 3, e69 (2005)
McGuinness, B. E., Hirota, T., Kudo, N. R., Peters, J. M. & Nasmyth, K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 3, e86 (2005)
Rigaut, G. et al. A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol. 17, 1030–1032 (1999)
Kinoshita, N., Yamano, H., Niwa, H., Yoshida, T. & Yanagida, M. Negative regulation of mitosis by the fission yeast protein phosphatase ppa2. Genes Dev. 7, 1059–1071 (1993)
Kinoshita, N., Ohkura, H. & Yanagida, M. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell 63, 405–415 (1990)
Kinoshita, K. et al. The regulatory subunits of fission yeast protein phosphatase 2A (PP2A) affect cell morphogenesis, cell wall synthesis and cytokinesis. Genes Cells 1, 29–45 (1996)
Jiang, W. & Hallberg, R. L. Isolation and characterization of par1+ and par2+: two Schizosaccharomyces pombe genes encoding B′ subunits of protein phosphatase 2A. Genetics 154, 1025–1038 (2000)
Ronne, H., Carlberg, M., Hu, G. Z. & Nehlin, J. O. Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis. Mol. Cell. Biol. 11, 4876–4884 (1991)
Zhao, Y., Boguslawski, G., Zitomer, R. S. & DePaoli-Roach, A. A. Saccharomyces cerevisiae homologs of mammalian B and B′ subunits of protein phosphatase 2A direct the enzyme to distinct cellular functions. J. Biol. Chem. 272, 8256–8262 (1997)
Kittler, R. et al. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc. Natl Acad. Sci. USA 102, 2396–2401 (2005)
Lew, D. J. & Burke, D. J. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37, 251–282 (2003)
Izawa, D., Goto, M., Yamashita, A., Yamano, H. & Yamamoto, M. Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature 434, 529–533 (2005)
Shimoda, C., Hirata, A., Kishida, M., Hashida, T. & Tanaka, K. Characterization of meiosis-deficient mutants by electron microscopy and mapping of four essential genes in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 200, 252–257 (1985)
Katou, Y. et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078–1083 (2003)
Lengronne, A. et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430, 573–578 (2004)
Meluh, P. B. & Koshland, D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11, 3401–3412 (1997)
Meluh, P. B., Yang, P., Glowczewski, L., Koshland, D. & Smith, M. M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94, 607–613 (1998)
Glynn, E. F. et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2, e259 (2004)
Kitajima, T. S., Yokobayashi, S., Yamamoto, M. & Watanabe, Y. Distinct cohesin complexes organize meiotic chromosome domains. Science 300, 1152–1155 (2003)
Watanabe, Y. & Nurse, P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400, 461–464 (1999)
Angell, R. First-meiotic-division nondisjunction in human oocytes. Am. J. Hum. Genet. 61, 23–32 (1997)
Kuliev, A., Cieslak, J., Ilkevitch, Y. & Verlinsky, Y. Chromosomal abnormalities in a series of 6,733 human oocytes in preimplantation diagnosis for age-related aneuploidies. Reprod. Biomed. Online 6, 54–59 (2003)
Pellestor, F., Andreo, B., Arnal, F., Humeau, C. & Demaille, J. Maternal aging and chromosomal abnormalities: new data drawn from in vitro unfertilized human oocytes. Hum. Genet. 112, 195–203 (2003)
Mailhes, J. B., Hilliard, C., Fuseler, J. W. & London, S. N. Okadaic acid, an inhibitor of protein phosphatase 1 and 2A, induces premature separation of sister chromatids during meiosis I and aneuploidy in mouse oocytes in vitro. Chromosome Res. 11, 619–631 (2003)
Yokobayashi, S., Yamamoto, M. & Watanabe, Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol. Cell. Biol. 23, 3965–3973 (2003)
Alexandru, G., Uhlmann, F., Mechtler, K., Poupart, M. A. & Nasmyth, K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105, 459–472 (2001)
Clyne, R. K. et al. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nature Cell Biol. 5, 480–485 (2003)
Lee, B. H. & Amon, A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300, 482–486 (2003)
Yu, H. G. & Koshland, D. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell 123, 397–407 (2005)
McCright, B., Rivers, A. M., Audlin, S. & Virshup, D. M. The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J. Biol. Chem. 271, 22081–22089 (1996)
Puig, O. et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 (2001)
Peters, A. H., Plug, A. W., van Vugt, M. J. & de Boer, P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 (1997)
Pidoux, A., Mellone, B. & Allshire, R. Analysis of chromatin in fission yeast. Methods 33, 252–259 (2004)
Yamamoto, M. The molecular control mechanisms of meiosis in fission yeast. Trends Biochem. Sci. 21, 18–22 (1996)
Acknowledgements
The authors would like to thank K. Rabitsch, N. Kudo, C. Kraft, I. Steinmacher, R. Imre, K. Tanaka, R. Kittler, F. Stewart and A. Lorenz for help with experiments, as well as M. Yanagida, I. Poser, I. Hagan, K. Gull, K. Gould, C. Warren, F. Spencer, D. Virshup, W. Zachariae and Y. Watanabe for cell lines, strains and reagents. In addition we would like to thank members of the Nasmyth laboratory for discussions. J.G. was a recipient of the EMBO long-term fellowship. K.M. received funding from GenAU. E.O. was supported by a grant from the Austrian Science Foundation. Author Contributions Experiments were designed and data analysed and interpreted by C.R., V.K. and K.N. S. pombe strain construction, cytological and biochemical analysis, TAP purification, conventional ChIP experiments and in vitro binding assays were performed by C.R. S. cerevisiae strain construction and cytological analysis was performed by V.K. ChIP using S. pombe and S. cerevisiae oligonucleotide microarrays was performed by Y.K., S.M., T.I. and K.S. S. cerevisiae TAP purification was performed by M.P. HeLa TAP purification was performed by J.G. from a cell line with help from F.B. and L.P. Cytological analysis of HeLa and NIH3T3 cells was performed by B.C., using cell lines generated by I.M. and E.O. Generation and characterization of human αB56 antibody was performed by I.M. and E.O. Mass spectrometry was performed by K.M. The manuscript was written by V.K., C.R. and K.N. W.H. and M.G. provided technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Methods, Supplementary Tables 1 and 2, and Supplementary Figures 1–3, 6–10 and Supplementary Figure Legends. (PDF 5310 kb)
Supplementary Figure 4
Distribution of Sp_Rec8, Sp_Par1B’, Sp_Ppa2C, and Sp_Sgo1 on chromatin, using a high-density oligonucleotide array of Schizosaccharomyces pombe chromosome 2 and most of chromosome 3. (PDF 9838 kb)
Supplementary Figure 5
Distribution of Sp_Par1B’ on chromatin in Sp_sgo1Δ cells, using a high-density oligonucleotide array of S. pombe chromosome 2 and most of chromosome 3. (PDF 3872 kb)
Rights and permissions
About this article
Cite this article
Riedel, C., Katis, V., Katou, Y. et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441, 53–61 (2006). https://doi.org/10.1038/nature04664
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04664
This article is cited by
-
Genome control by SMC complexes
Nature Reviews Molecular Cell Biology (2023)
-
SGOL2 is a novel prognostic marker and fosters disease progression via a MAD2-mediated pathway in hepatocellular carcinoma
Biomarker Research (2022)
-
ATM phosphorylates PP2A subunit A resulting in nuclear export and spatiotemporal regulation of the DNA damage response
Cellular and Molecular Life Sciences (2022)
-
Genomewide expression analysis of the heat stress response in dermal fibroblasts of Tharparkar (zebu) and Karan-Fries (zebu × taurine) cattle
Cell Stress and Chaperones (2020)
-
Construction of strains to identify novel factors for regulation of centromeric cohesion protection (CCP) and sister kinetochore mono-orientation (SKM)
BMC Molecular and Cell Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.