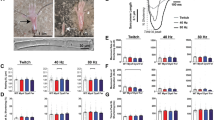
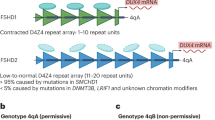
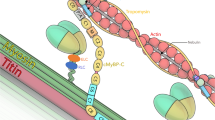
Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant neuromuscular disorder that is not due to a classical mutation within a protein-coding gene1,2. Instead, almost all FSHD patients carry deletions of an integral number of tandem 3.3-kilobase repeat units, termed D4Z4, located on chromosome 4q35 (ref. 3). D4Z4 contains a transcriptional silencer whose deletion leads to inappropriate overexpression in FSHD skeletal muscle of 4q35 genes located upstream of D4Z4 (ref. 4). To identify the gene responsible for FSHD pathogenesis, we generated transgenic mice selectively overexpressing in skeletal muscle the 4q35 genes FRG1, FRG2 or ANT1. We find that FRG1 transgenic mice develop a muscular dystrophy with features characteristic of the human disease; by contrast, FRG2 and ANT1 transgenic mice seem normal. FRG1 is a nuclear protein and several lines of evidence suggest it is involved in pre-messenger RNA splicing5,6,7. We find that in muscle of FRG1 transgenic mice and FSHD patients, specific pre-mRNAs undergo aberrant alternative splicing. Collectively, our results suggest that FSHD results from inappropriate overexpression of FRG1 in skeletal muscle, which leads to abnormal alternative splicing of specific pre-mRNAs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tawil, R. Facioscapulohumeral muscular dystrophy. Curr. Neurol. Neurosci. Rep. 4, 51–54 (2004)
Flanigan, K. M. in Myology (eds Engel, A. & Franzini-Armstrong, C.) 1123–1133 (McGraw Hill Professional, New York, 2004)
van Deutekom, J. C. et al. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 2, 2037–2042 (1993)
Gabellini, D., Green, M. R. & Tupler, R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110, 339–348 (2002)
Kim, S. K. et al. A gene expression map for Caenorhabditis elegans. Science 293, 2087–2092 (2001)
Rappsilber, J., Ryder, U., Lamond, A. I. & Mann, M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12, 1231–1245 (2002)
van Koningsbruggen, S. et al. FRG1P is localised in the nucleolus, Cajal bodies, and speckles. J. Med. Genet. 41, e46 (2004)
Muscat, G. E. & Kedes, L. Multiple 5′-flanking regions of the human α-skeletal actin gene synergistically modulate muscle-specific expression. Mol. Cell. Biol. 7, 4089–4099 (1987)
Ricci, E. et al. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann. Neurol. 45, 751–757 (1999)
Kilmer, D. D. et al. Profiles of neuromuscular diseases. Facioscapulohumeral muscular dystrophy. Am. J. Phys. Med. Rehabil. 74, S131–S139 (1995)
Deconinck, A. E. et al. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell 90, 717–727 (1997)
Grady, R. M. et al. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell 90, 729–738 (1997)
Lynch, G. S., Hinkle, R. T., Chamberlain, J. S., Brooks, S. V. & Faulkner, J. A. Force and power output of fast and slow skeletal muscles from mdx mice 6–28 months old. J. Physiol. (Lond.) 535, 591–600 (2001)
Durbeej, M. & Campbell, K. P. Muscular dystrophies involving the dystrophin–glycoprotein complex: an overview of current mouse models. Curr. Opin. Genet. Dev. 12, 349–361 (2002)
Muntoni, F., Brockington, M. & Brown, S. C. Glycosylation eases muscular dystrophy. Nature Med. 10, 676–677 (2004)
Dalkilic, I. & Kunkel, L. M. Muscular dystrophies: genes to pathogenesis. Curr. Opin. Genet. Dev. 13, 231–238 (2003)
Calado, A. et al. Nuclear inclusions in oculopharyngeal muscular dystrophy consist of poly(A) binding protein 2 aggregates which sequester poly(A) RNA. Hum. Mol. Genet. 9, 2321–2328 (2000)
Gubitz, A. K., Feng, W. & Dreyfuss, G. The SMN complex. Exp. Cell Res. 296, 51–56 (2004)
Ranum, L. P. & Day, J. W. Myotonic dystrophy: RNA pathogenesis comes into focus. Am. J. Hum. Genet. 74, 793–804 (2004)
Buj-Bello, A. et al. Muscle-specific alternative splicing of myotubularin-related 1 gene is impaired in DM1 muscle cells. Hum. Mol. Genet. 11, 2297–2307 (2002)
Kanadia, R. N. et al. A muscleblind knockout model for myotonic dystrophy. Science 302, 1978–1980 (2003)
Orrell, R. W. et al. Definitive molecular diagnosis of facioscapulohumeral dystrophy. Neurology 52, 1822–1826 (1999)
Rogers, M., Sewry, C. A. & Upadhyaya, M. in FSHD Facioscapulohumeral Muscular Dystrophy: Clinical Medicine and Molecular Cell Biology (eds Upadhyaya, M. & Cooper, D. N.) 277–298 (BIOS Scientific Publishers, London and New York, 2004)
Lemmers, R. J. et al. D4F104S1 deletion in facioscapulohumeral muscular dystrophy: phenotype, size, and detection. Neurology 61, 178–183 (2003)
Tupler, R. et al. Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J. Med. Genet. 33, 366–370 (1996)
Chaudhuri, T., Mukherjea, M., Sachdev, S., Randall, J. D. & Sarkar, S. Role of the fetal and α/β exons in the function of fast skeletal troponin T isoforms: correlation with altered Ca2+ regulation associated with development. J. Mol. Biol. 352, 58–71 (2005)
Johnson, J. M. et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302, 2141–2144 (2003)
Dubowitz, V. Muscle Biopsy. A Practical Approach (Bailliere Tindall, London, 1985)
Acknowledgements
We are indebted to all FSHD families and the FSH Society. We thank M. Mora for providing FSHD myoblasts; G. Di Giulio for performing the radiology; C. Ghigna, S. Jones, L. Castilla and members of the M.R.G. laboratory for helpful discussions; and S. Evans for assistance with preparing the manuscript. Space constraints have limited the amount of original work that we have been able to cover in this Letter. This work was supported by the Muscular Dystrophy Association (D.G.); Associazione Amici del Centro Dino Ferrari, Telethon Bank, Eurobiobank and R.F. 2002 Criobanca Automatizzata di Materiale Biologico (M.M.); Telethon (R.B.); and the Cariplo Fundation, NIH-NINDS, the Muscular Dystrophy Association, the Italian National Council for Research Progetto Genomica Funzionale, the Association Francaise contre les Myopaties, the Italian Ministry of Education, University, and Research, Legato Ferrari and Telethon (R.T). M.R.G. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Drs Gabellini, Tupler and Green have filed U.S. patent application 20050054012, entitled “Methods of detecting and treating FSHD”.
Supplementary information
Supplementary Methods
Methods for RT-PCR analysis, real time RT–PCR, FRG1 transgene copy number determination, FRG1 transgene northern blotting, histological and histochemical analyses, analysis of sarcolemmal integrity, immunofluorescence analysis of FRG1, overexpression of ASF/SF2 in C2C12 cells.
Supplementary Figures
This file contains Supplementary Figures 1–9.
Supplementary Tables 1 and 2
Supplementary Table 1 details the histopathological and histochemical comparison of ten different skeletal muscles of FRG1 transgenic mice. Supplementary Table 2 details the comparison of the muscles affected and the degree of dystrophy between FSHD patients and FRG1 transgenic mice.
Rights and permissions
About this article
Cite this article
Gabellini, D., D'Antona, G., Moggio, M. et al. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature 439, 973–977 (2006). https://doi.org/10.1038/nature04422
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04422
This article is cited by
-
Identification of a novel isolated 4q35.2 microdeletion in a Chinese pediatric patient using chromosomal microarray analysis: a case report and literature review
Molecular Cytogenetics (2023)
-
Transplantation of PSC-derived myogenic progenitors counteracts disease phenotypes in FSHD mice
npj Regenerative Medicine (2022)
-
Reduced FRG1 expression promotes prostate cancer progression and affects prostate cancer cell migration and invasion
BMC Cancer (2019)
-
Muscle pathology from stochastic low level DUX4 expression in an FSHD mouse model
Nature Communications (2017)
-
Genome engineering: a new approach to gene therapy for neuromuscular disorders
Nature Reviews Neurology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.