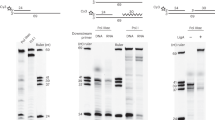
Abstract
Unrepaired lesions in the DNA template pose a threat to accurate replication. Several pathways exist in Escherichia coli to reactivate a blocked replication fork. The process of recombination-dependent restart of broken forks is well understood, but the consequence of replication through strand-specific lesions is less well known. Here we show that replication can be restarted and leading-strand synthesis re-initiated downstream of an unrepaired block to leading-strand progression, even when the 3′-OH of the nascent leading strand is unavailable. We demonstrate that the loading by a replication restart system of a single hexamer of the replication fork helicase, DnaB, on the lagging-strand template is sufficient to coordinate priming by the DnaG primase of both the leading and lagging strands. These observations provide a mechanism for damage bypass during fork reactivation, demonstrate how daughter-strand gaps are generated opposite leading-strand lesions during the replication of ultraviolet-light-irradiated DNA, and help to explain the remarkable speed at which even a heavily damaged DNA template is replicated.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Myung, K., Chen, C. & Kolodner, R. D. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411, 1073–1076 (2001)
Witkin, E. M. Ultraviolet-induced mutation and DNA repair. Annu. Rev. Microbiol. 23, 487–514 (1969)
Rupp, W. D. & Howard-Flanders, P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 31, 291–304 (1968)
Iyer, V. N. & Rupp, W. D. Usefulness of benzoylated naphthoylated DEAE-cellulose to distinguish and fractionate double-stranded DNA bearing different extents of single-stranded regions. Biochim. Biophys. Acta 228, 117–126 (1971)
Bridges, B. A. & Sedgwick, S. G. Effect of photoreactivation on the filling of gaps in deoxyribonucleic acid synthesized after exposure of Escherichia coli to ultraviolet light. J. Bacteriol. 117, 1077–1081 (1974)
Ganesan, A. K. Persistence of pyrimidine dimers during post-replication repair in ultraviolet light-irradiated Escherichia coli K12. J. Mol. Biol. 87, 103–119 (1974)
Bridges, B. A. & Munson, R. J. Mutagenesis in Escherichia coli: evidence for the mechanism of base change mutation by ultraviolet radiation in a strain deficient in excision-repair. Proc. R. Soc. Lond. B 171, 213–226 (1968)
Higuchi, K. et al. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells 8, 437–449 (2003)
McInerney, P. & O'Donnell, M. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 279, 21543–21551 (2004)
Pages, V. & Fuchs, R. P. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science 300, 1300–1303 (2003)
Higgins, N. P., Kato, K. & Strauss, B. A model for replication repair in mammalian cells. J. Mol. Biol. 101, 417–425 (1976)
Seigneur, M., Bidnenko, V., Ehrlich, S. D. & Michel, B. RuvAB acts at arrested replication forks. Cell 95, 419–430 (1998)
Sandler, S. J. & Marians, K. J. Role of PriA in replication fork reactivation in Escherichia coli. J. Bacteriol. 182, 9–13 (2000)
Michel, B., Grompone, G., Flores, M. J. & Bidnenko, V. Multiple pathways process stalled replication forks. Proc. Natl Acad. Sci. USA 101, 12783–12788 (2004)
Heller, R. C. & Marians, K. J. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol. Cell 17, 733–743 (2005)
Wu, C. A., Zechner, E. L. & Marians, K. J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. I. Multiple effectors act to modulate Okazaki fragment size. J. Biol. Chem. 267, 4030–4044 (1992)
Xu, L. & Marians, K. J. PriA mediates DNA replication pathway choice at recombination intermediates. Mol. Cell 11, 817–826 (2003)
Swart, J. R. & Griep, M. A. Primase from Escherichia coli primes single-stranded templates in the absence of single-stranded DNA-binding protein or other auxiliary proteins. Template sequence requirements based on the bacteriophage G4 complementary strand origin and Okazaki fragment initiation sites. J. Biol. Chem. 268, 12970–12976 (1993)
Tougu, K., Peng, H. & Marians, K. J. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J. Biol. Chem. 269, 4675–4682 (1994)
Hacker, K. J. & Johnson, K. A. A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry 36, 14080–14087 (1997)
Kaplan, D. L. & O'Donnell, M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell 10, 647–657 (2002)
Xu, L. & Marians, K. J. Purification and characterization of DnaC810, a primosomal protein capable of bypassing PriA function. J. Biol. Chem. 275, 8196–8205 (2000)
Galletto, R., Jezewska, M. J. & Bujalowski, W. Interactions of the Escherichia coli DnaB helicase hexamer with the replication factor the DnaC protein. Effect of nucleotide cofactors and the ssDNA on protein-protein interactions and the topology of the complex. J. Mol. Biol. 329, 441–465 (2003)
Davey, M. J., Fang, L., McInerney, P., Georgescu, R. E. & O'Donnell, M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 21, 3148–3159 (2002)
Reha-Krantz, L. J. & Hurwitz, J. The dnaB gene product of Escherichia coli. II. Single stranded DNA-dependent ribonucleoside triphosphatase activity. J. Biol. Chem. 253, 4051–4057 (1978)
Mitkova, A. V., Khopde, S. M. & Biswas, S. B. Mechanism and stoichiometry of interaction of DnaG primase with DnaB helicase of Escherichia coli in RNA primer synthesis. J. Biol. Chem. 278, 52253–52261 (2003)
Smith, K. C. Recombinational DNA repair: the ignored repair systems. Bioessays 26, 1322–1326 (2004)
Grompone, G., Sanchez, N., Dusko Ehrlich, S. & Michel, B. Requirement for RecFOR-mediated recombination in priA mutant. Mol. Microbiol. 52, 551–562 (2004)
Sandler, S. J. Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics 155, 487–497 (2000)
Prakash, L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 184, 471–478 (1981)
Lehmann, A. R. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J. Mol. Biol. 66, 319–337 (1972)
Meneghini, R. Gaps in DNA synthesized by ultraviolet light-irradiated WI38 human cells. Biochim. Biophys. Acta 425, 419–427 (1976)
Okazaki, R., Okazaki, T., Sakabe, K., Sugimoto, K. & Sugino, A. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc. Natl Acad. Sci. USA 59, 598–605 (1968)
Ogawa, T. & Okazaki, T. Discontinuous DNA replication. Annu. Rev. Biochem. 49, 421–457 (1980)
Acknowledgements
These studies were supported by a grant from the NIH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Heller, R., Marians, K. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439, 557–562 (2006). https://doi.org/10.1038/nature04329
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04329
This article is cited by
-
Leveraging the replication stress response to optimize cancer therapy
Nature Reviews Cancer (2023)
-
Arabidopsis thaliana PrimPol is a primase and lesion bypass DNA polymerase with the biochemical characteristics to cope with DNA damage in the nucleus, mitochondria, and chloroplast
Scientific Reports (2021)
-
DNA replication stress: from molecular mechanisms to human disease
Chromosoma (2017)
-
Replication stress: getting back on track
Nature Structural & Molecular Biology (2016)
-
Transcription–replication conflicts: how they occur and how they are resolved
Nature Reviews Molecular Cell Biology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.