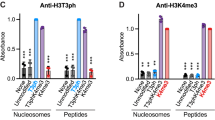
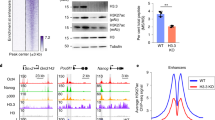
Abstract
Tri-methylation of histone H3 lysine 9 is important for recruiting heterochromatin protein 1 (HP1) to discrete regions of the genome, thereby regulating gene expression, chromatin packaging and heterochromatin formation. Here we show that HP1α, -β, and -γ are released from chromatin during the M phase of the cell cycle, even though tri-methylation levels of histone H3 lysine 9 remain unchanged. However, the additional, transient modification of histone H3 by phosphorylation of serine 10 next to the more stable methyl-lysine 9 mark is sufficient to eject HP1 proteins from their binding sites. Inhibition or depletion of the mitotic kinase Aurora B, which phosphorylates serine 10 on histone H3, causes retention of HP1 proteins on mitotic chromosomes, suggesting that H3 serine 10 phosphorylation is necessary for the dissociation of HP1 from chromatin in M phase. These findings establish a regulatory mechanism of protein–protein interactions, through a combinatorial readout of two adjacent post-translational modifications: a stable methylation and a dynamic phosphorylation mark.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Felsenfeld, G. & Groudine, M. Controlling the double helix. Nature 421, 448–453 (2003)
Khorasanizadeh, S. The nucleosome: from genomic organization to genomic regulation. Cell 116, 259–272 (2004)
Grewal, S. I. & Moazed, D. Heterochromatin and epigenetic control of gene expression. Science 301, 798–802 (2003)
Grewal, S. I. & Elgin, S. C. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12, 178–187 (2002)
Li, Y., Kirschmann, D. A. & Wallrath, L. L. Does heterochromatin protein 1 always follow code? Proc. Natl Acad. Sci. USA 99 (suppl.), 16462–16469 (2002)
Eissenberg, J. C. & Elgin, S. C. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10, 204–210 (2000)
Minc, E., Allory, Y., Worman, H. J., Courvalin, J. C. & Buendia, B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108, 220–234 (1999)
Hayakawa, T., Haraguchi, T., Masumoto, H. & Hiraoka, Y. Cell cycle behaviour of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J. Cell Sci. 116, 3327–3338 (2003)
Schmiedeberg, L., Weisshart, K., Diekmann, S., Meyer Zu Hoerste, G. & Hemmerich, P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol. Biol. Cell 15, 2819–2833 (2004)
Pidoux, A. L. & Allshire, R. C. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 12, 521–534 (2004)
Stewart, M. D., Li, J. & Wong, J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol. Cell. Biol. 25, 2525–2538 (2005)
Peters, A. H. et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 (2003)
Thiru, A. et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 23, 489–499 (2004)
Sims, R. J. III, Nishioka, K. & Reinberg, D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 19, 629–639 (2003)
Schotta, G., Lachner, M., Peters, A. H. & Jenuwein, T. The indexing potential of histone lysine methylation. Novartis Found. Symp. 259, 22–37 (2004)
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001)
Jacobs, S. A. et al. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20, 5232–5241 (2001)
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001)
Jacobs, S. A. & Khorasanizadeh, S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295, 2080–2083 (2002)
Fischle, W. et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17, 1870–1881 (2003)
Maison, C. & Almouzni, G. HP1 and the dynamics of heterochromatin maintenance. Nature Rev. Mol. Cell Biol. 5, 296–304 (2004)
Cheutin, T. et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299, 721–725 (2003)
Festenstein, R. et al. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299, 719–721 (2003)
Maison, C. et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nature Genet. 30, 329–334 (2002)
Mateescu, B., England, P., Halgand, F., Yaniv, M. & Muchardt, C. Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep. 5, 490–496 (2004)
Syka, J. E. et al. Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J. Proteome Res. 3, 621–626 (2004)
Hendzel, M. J. et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106, 348–360 (1997)
Prigent, C. & Dimitrov, S. Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. 116, 3677–3685 (2003)
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000)
Yasui, Y. et al. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J. Biol. Chem. 279, 12997–13003 (2004)
Honda, R., Korner, R. & Nigg, E. A. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell 14, 3325–3341 (2003)
Chen, J. et al. Survivin enhances Aurora-B kinase activity and localizes Aurora-B in human cells. J. Biol. Chem. 278, 486–490 (2003)
Murray, A. W. Cell cycle extracts. Methods Cell Biol. 36, 581–605 (1991)
Andrews, P. D., Knatko, E., Moore, W. J. & Swedlow, J. R. Mitotic mechanics: the auroras come into view. Curr. Opin. Cell Biol. 15, 672–683 (2003)
Hauf, S. et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281–294 (2003)
Sampath, S. C. et al. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118, 187–202 (2004)
Nielsen, P. R. et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416, 103–107 (2002)
Sugimoto, K., Tasaka, H. & Dotsu, M. Molecular behaviour in living mitotic cells of human centromere heterochromatin protein HPLα ectopically expressed as a fusion to red fluorescent protein. Cell Struct. Funct. 26, 705–718 (2001)
Fischle, W., Wang, Y. & Allis, C. D. Binary switches and modification cassettes in histone biology and beyond. Nature 425, 475–479 (2003)
Obuse, C. et al. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nature Cell Biol. 6, 1135–1141 (2004)
Ditchfield, C. et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, 267–280 (2003)
Lampson, M. A., Renduchitala, K., Khodjakov, A. & Kapoor, T. M. Correcting improper chromosome–spindle attachments during cell division. Nature Cell Biol. 6, 232–237 (2004)
Mellone, B. G. et al. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr. Biol. 13, 1748–1757 (2003)
Wei, Y., Yu, L., Bowen, J., Gorovsky, M. A. & Allis, C. D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97, 99–109 (1999)
Hsu, J. Y. et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 (2000)
Dai, J., Sultan, S., Taylor, S. S. & Higgins, J. M. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19, 472–488 (2005)
Jacobs, S. A., Fischle, W. & Khorasanizadeh, S. Assays for the determination of structure and dynamics of the interaction of the chromodomain with histone peptides. Methods Enzymol. 376, 131–148 (2004)
Acknowledgements
We are indebted to M. A. Jelinek and colleagues at Upstate Biotechnologies for developing the monoclonal dual-mark combination-specific anti-H3K9me3S10ph antibody, to S. Hake and C. Barber for purifying H3 for mass spectrometry analysis, and to S. Mollah for initial mass spectrometry analyses. We thank T. Kapoor and Boehringer Ingelheim for providing hesperadin, S. Taylor for the anti-Aurora B antibody, and P. Hemmerich for the HP1–GFP expression constructs. We are grateful to S. Khorasanizadeh and S. Jacobs for their input and help with intepretation of structural data, and to S. Sampath and E. Zeleneova for their input at early stages of this work. This work was funded by grants from the National Institutes of Health (C.D.A. and D.H.F.) and by The Rockefeller University (C.D.A. and H.F.). H.F. is supported by a Searle Scholarship, the Alexandrine and Alexander Sinsheimer Fund, and the Irma T.Hirschl/Monique Weill-Caulier Trust. W.F. is a Robert Black fellow of the Damon Runyon Cancer Research Foundation. H.L.D. is supported by a predoctoral fellowship from the Boehringer Ingelheim Foundation and B.S.T. is supported by an NRSA Training Grant.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure S1
Characterization of the monoclonal anti-H3K9me3S10ph antibody in various applications. (PDF 823 kb)
Supplementary Figure S2
Sequence and structural comparison of HP1 isoforms from different organisms. Binding data of full-length HP1 and HP1β mutant proteins to methylated H3 peptides. (PDF 709 kb)
Supplementary Figure S3
Quantitative phosphorylation analysis of H3-tail peptides methylated on K9. The Aurora B kinase containing chromosomal passenger complex (CPC) phosphorylates H3S10 independent of the methylation status of H3K9. (PDF 32 kb)
Supplementary Figure S4
Phosphorylation of H3K9me3 in the presence of increasing amounts of different HP1 isoforms. Fluorescence polarization analysis of the HP1α, β, and γ isoforms shows decreased binding to H3K9me3 after phosphorylation by the chromosomal passenger complex (CPC). (PDF 1524 kb)
Supplementary Figure S5
Metaphase spreads showing the distribution of the dual-mark combination of H3K9me3S10ph on M-phase chromosomes. (PDF 292 kb)
Supplementary Figure S6
Immunofluorescence analysis of 10T1/2 cells with anti-H3K9me3S10ph and anti-HP1α, β, and γ antibodies in the absence or presence of the Aurora B kinase inhibitor hesperadin (Field shot and individual Interphase cells). (PDF 590 kb)
Supplementary Figure S7
Different distribution of GFP-HP1α, β, and γ in mitotic HeP-2 cells untreated or treated with the Aurora B inhibitor hesperadin. Inhibition of mitotic H3S10 phosphorylation results in aberrant association of exogenous HP1 with M-phase chromatin. (PDF 408 kb)
Supplementary Figure S8
Effect of Aurora B knock-down on H3K9me3S10ph and HP1 distribution in M-phase. Absence of Aurora B leads to aberrant association of endogenous HP1 with M-phase chromatin. (PDF 323 kb)
Supplementary Figure S9
Immunofluorescence analysis of chromosomes reconstituted in Xenopus egg extracts shows the conservation of the dual-mark of H3K9me3S10ph and the specificity of the monoclonal anti-H3K9me3S10ph antibody. (PDF 65 kb)
Supplementary Figure S10
Depletion of the chromosomal passenger complex (CPC) does not affect methylation of H3K9 in Xenopus egg extracts. (PDF 41 kb)
Supplementary Figure S11
Characterization of the anti-xHP1α antibodies. (PDF 37 kb)
Supplementary Table S1
Antibodies and dilutions used in study. (RTF 42 kb)
Supplementary Figure Legends
Figure legends to Supplementary Figures S1–S11. (RTF 19 kb)
Supplementary Methods
Detailed description of experimental methods used in study. (RTF 23 kb)
Supplementary Notes
Bibliography of references cited in Supplementary Information. (RTF 6 kb)
Rights and permissions
About this article
Cite this article
Fischle, W., Tseng, B., Dormann, H. et al. Regulation of HP1–chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 (2005). https://doi.org/10.1038/nature04219
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04219
This article is cited by
-
Orchestrating chromosome conformation capture analysis with Bioconductor
Nature Communications (2024)
-
EOAI, a ubiquitin-specific peptidase 5 inhibitor, prevents non-small cell lung cancer progression by inducing DNA damage
BMC Cancer (2023)
-
Release of Histone H3K4-reading transcription factors from chromosomes in mitosis is independent of adjacent H3 phosphorylation
Nature Communications (2023)
-
TNRC18 engages H3K9me3 to mediate silencing of endogenous retrotransposons
Nature (2023)
-
USP8 inhibitor–induced DNA damage activates cell cycle arrest, apoptosis, and autophagy in esophageal squamous cell carcinoma
Cell Biology and Toxicology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.