Abstract
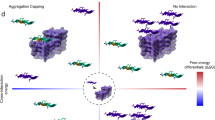
Incorrect folding of proteins, leading to aggregation and amyloid formation, is associated with a group of highly debilitating medical conditions1,2 including Alzheimer's disease and late-onset diabetes. The issue of how unwanted protein association is normally avoided in a living system is particularly significant in the context of the evolution of multidomain proteins, which account for over 70% of all eukaryotic proteins3, where the effective local protein concentration in the vicinity of each domain is very high. Here we describe the aggregation kinetics of multidomain protein constructs of immunoglobulin domains and the ability of different homologous domains to aggregate together. We show that aggregation of these proteins is a specific process and that the efficiency of coaggregation between different domains decreases markedly with decreasing sequence identity. Thus, whereas immunoglobulin domains with more than about 70% identity are highly prone to coaggregation, those with less than 30–40% sequence identity do not detectably interact. A bioinformatics analysis of consecutive homologous domains in large multidomain proteins shows that such domains almost exclusively have sequence identities of less than 40%, in other words below the level at which coaggregation is likely to be efficient. We propose that such low sequence identities could have a crucial and general role in safeguarding proteins against misfolding and aggregation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Dobson, C. M. Protein folding and misfolding. Nature 426, 884–890 (2003)
Selkoe, D. J. Folding proteins in fatal ways. Nature 426, 900–904 (2003)
Apic, G., Gough, J. & Teichmann, S. A. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 310, 311–325 (2001)
Biere, A. L. et al. Parkinson's disease-associated α-synuclein is more fibrillogenic than β- and γ-synuclein and cannot cross-seed its homologs. J. Biol. Chem. 275, 34574–34579 (2000)
Krebs, M. R. H., Morozova-Roche, L. A., Daniel, K., Robinson, C. V. & Dobson, C. M. Observation of sequence specificity in the seeding of protein amyloid fibrils. Protein Sci. 13, 1933–1938 (2004)
Silow, M., Tan, Y.-J., Fersht, A. R. & Oliveberg, M. Formation of short-lived protein aggregates directly from the coil in two-state folding. Biochemistry 38, 13006–13012 (1999)
Labeit, S. et al. A regular pattern of two types of 100-residue motif in the sequence of titin. Nature 345, 273–276 (1990)
Linke, W. A., Stockmeier, M. R., Ivemeyer, M., Hosser, H. & Mundel, P. Characterizing titin's I-band Ig domain region as an entropic spring. J. Cell Sci. 111, 1567–1574 (1998)
Raffen, R. et al. Physicochemical consequences of amino acid variations that contribute to fibril formation by immunoglobulin light chains. Protein Sci. 8, 509–517 (1999)
McParland, V. J. et al. Partially unfolded states of β2-microglobulin and amyloid formation in vitro. Biochemistry 39, 8735–8746 (2000)
Oberhauser, A. F., Marszalek, P. E., Carrion-Vasquez, M. & Fernandez, A. Single protein misfolding events captured by atomic force microscopy. Nature Struct. Biol. 6, 1025–1028 (1999)
Chiti, F. et al. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl Acad. Sci. USA 96, 3590–3594 (1999)
Sunde, M. et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 273, 729–739 (1997)
Oosawa, F. & Asakura, S. Thermodynamics of the Polymerization of Protein (Academic, London, 1975)
Steward, A., Toca-Herrera, J. L. & Clarke, J. Versatile cloning system for construction of multimeric proteins for use in atomic force microscopy. Protein Sci. 11, 2179–2183 (2002)
Scott, K. A., Steward, A., Fowler, S. B. & Clarke, J. Titin: a multidomain protein that behaves as the sum of its parts. J. Mol. Biol. 315, 819–829 (2002)
Ellis, R. J. & Minton, A. P. Join the crowd. Nature 425, 27–28 (2003)
Pepys, M. B. Pathogenesis, diagnosis and treatment of systemic amyloidosis. Phil. Trans. R. Soc. Lond. B 356, 203–211 (2001)
Rajan, R. S., Illing, M. E., Bence, N. F. & Kopito, R. Specificity in intracellular protein aggregation and inclusion body formation. Proc. Natl Acad. Sci. USA 98, 13060–13065 (2001)
Nelson, R. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005)
Jaroniec, C. P. et al. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc. Natl Acad. Sci. USA 101, 711–716 (2004)
Krishnan, R. & Lindquist, S. L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 (2005)
Otzen, D. E., Kristensen, O. & Oliveberg, M. Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: a structural clue to amyloid assembly. Proc. Natl Acad. Sci. USA 97, 9907–9912 (2000)
Steward, A., Adhya, S. & Clarke, J. Sequence conservation in Ig-like domains: the role of highly conserved proline residues in the fibronectin type III superfamily. J. Mol. Biol. 318, 935–940 (2002)
Richardson, J. S. & Richardson, D. C. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl Acad. Sci. USA 99, 2754–2759 (2002)
Netzer, W. J. & Hartl, F. U. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388, 343–349 (1997)
Barral, J. M., Broadley, S. A., Schaffar, G. F. & Hartl, F. U. Roles of molecular chaperones in protein misfolding diseases. Semin. Cell. Dev. Biol. 15, 17–29 (2004)
Goldberg, A. L. Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899 (2003)
Rutherford, S. L. & Lindquist, S. Hsp90 as a capacitor for morphological evolution. Nature 396, 336–342 (1998)
Gough, J. & Chothia, C. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 30, 268–272 (2002)
Acknowledgements
We thank K. Scott, A. Steward and J. Zurdo for clones; and R. Bader, D. Hall, C. Ponting and C. Chothia for discussions. This work was supported by the Wellcome Trust and the Leverhulme Trust (C.M.D.). C.F.W. held a Wellcome Trust prize studentship and J.C. is a Senior Wellcome Trust research fellow.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Figures 1–5, Supplementary Tables 1–5 and additional references. (DOC 4082 kb)
Rights and permissions
About this article
Cite this article
Wright, C., Teichmann, S., Clarke, J. et al. The importance of sequence diversity in the aggregation and evolution of proteins. Nature 438, 878–881 (2005). https://doi.org/10.1038/nature04195
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04195
This article is cited by
-
Agglomeration: when folded proteins clump together
Biophysical Reviews (2023)
-
The role of single-protein elasticity in mechanobiology
Nature Reviews Materials (2022)
-
Designed folding pathway of modular coiled-coil-based proteins
Nature Communications (2021)
-
Intrinsic disorder in protein domains contributes to both organism complexity and clade-specific functions
Scientific Reports (2021)
-
Mechanisms and therapeutic potential of interactions between human amyloids and viruses
Cellular and Molecular Life Sciences (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.