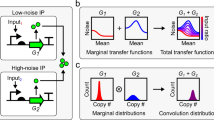
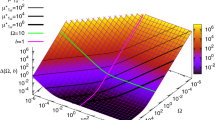
Abstract
Recent work demonstrates that stochastic fluctuations in molecular populations have consequences for gene regulation1,2,3,4,5,6,7,8,9,10. Previous experiments focused on noise sources or noise propagation through gene networks by measuring noise magnitudes. However, in theoretical analysis, we showed that noise frequency content is determined by the underlying gene circuits, leading to a mapping between gene circuit structure and the noise frequency range11,12. An intriguing prediction from our previous studies was that negative autoregulation shifts noise to higher frequencies where it is more easily filtered out by gene networks11—a property that may contribute to the prevalence of autoregulation motifs (for example, found in the regulation of ∼40% of Escherichia coli genes). Here we measure noise frequency content in growing cultures of E. coli, and verify the link between gene circuit structure and noise spectra by demonstrating the negative autoregulation-mediated spectral shift. We further demonstrate that noise spectral measurements provide mechanistic insights into gene regulation, as perturbations of gene circuit parameters are discernible in the measured noise frequency ranges. These results suggest that noise spectral measurements could facilitate the discovery of novel regulatory relationships.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002)
Rosenfeld, N., Young, J. W., Alon, U., Swain, P. S. & Elowitz, M. B. Gene regulation at the single-cell level. Science 307, 1962–1965 (2005)
Swain, P. S., Elowitz, M. B. & Siggia, E. D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl Acad. Sci. USA 99, 12795–12800 (2002)
Blake, W. J., Kaern, M., Cantor, C. R. & Collins, J. J. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003)
Raser, J. M. & O'Shea, E. K. Control of stochasticity in eukaryotic gene expression. Science 304, 1811–1814 (2004)
Ozbudak, E. M., Thattai, M., Kurtser, I., Grossman, A. D. & van Oudenaarden, A. Regulation of noise in the expression of a single gene. Nature Genet. 31, 69–73 (2002)
Pedraza, J. M. & van Oudenaarden, A. Noise propagation in gene networks. Science 307, 1965–1969 (2005)
Thattai, M. & van Oudenaarden, A. Intrinsic noise in gene regulatory networks. Proc. Natl Acad. Sci. USA 98, 8614–8619 (2001)
Kaern, M., Elston, T. C., Blake, W. J. & Collins, J. J. Stochasticity in gene expression: From theories to phenotypes. Nature Rev. Genet. 6, 451–464 (2005)
Rao, C. V., Wolf, D. M. & Arkin, A. P. Control, exploitation and tolerance of intracellular noise. Nature 420, 231–237 (2002)
Simpson, M. L., Cox, C. D. & Sayler, G. S. Frequency domain analysis of noise in autoregulated gene circuits. Proc. Natl Acad. Sci. USA 100, 4551–4556 (2003)
Simpson, M. L., Cox, C. D. & Sayler, G. S. Frequency domain chemical Langevin analysis of stochasticity in gene transcriptional regulation. J. Theor. Biol. 229, 383–394 (2004)
Gillespie, D. T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 (1977)
Gibson, M. A. & Bruck, J. Efficient exact stochastic simulation of chemical systems with many species and many channels. J. Phys. Chem. A 104, 1876–1889 (2000)
Andersen, J. B. et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64, 2240–2246 (1998)
Kepler, T. B. & Elston, T. C. Stochasticity in transcriptional regulation: Origins, consequences, and mathematical representations. Biophys. J. 81, 3116–3136 (2001)
Cox, C. D. et al. Analysis of noise in quorum sensing. OMICS 7, 317–334 (2003)
Lutz, R. & Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 (1997)
Becskei, A. & Serrano, L. Engineering stability in gene networks by autoregulation. Nature 405, 590–593 (2000)
Arkin, A., Ross, J. & McAdams, H. H. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics 149, 1633–1648 (1998)
Thattai, M. & van Oudenaarden, A. Stochastic gene expression in fluctuating environments. Genetics 167, 523–530 (2004)
Becskei, A., Seraphin, B. & Serrano, L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 20, 2528–2535 (2001)
Isaacs, F. J., Hasty, J., Cantor, C. R. & Collins, J. J. Prediction and measurement of an autoregulatory genetic module. Proc. Natl Acad. Sci. USA 100, 7714–7719 (2003)
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000)
Bendat, J. S. & Piersol, A. G. Data: Analysis and Measurement Procedures (Wiley, New York, 2000)
Acknowledgements
We thank I. Golding and J. Dunlap for advice on sample preparation and imaging, and M. Elowitz, J. Collins and T. Gardner for the gift of plasmids. This work was supported by the National Academies Keck Futures Initiative, the DARPA Bio-Computation Program, the NSF, the DOE Office of Advanced Scientific Computing Research, and was a user project of the Oak Ridge National Laboratory (ORNL) Center for Nanophase Materials Sciences (CNMS). D.W.A. acknowledges support from an ORNL CNMS Research Scholar Fellowship. R.D.D. acknowledges support from the DOE Science Undergraduate Laboratory Internship program. Author Contributions D.W.A., M.S.A., J.M.M., C.D.C. and M.L.S. planned the experimental, analytical and computational work. D.W.A., R.D.D., and J.R.W. performed the time-lapse microscopy measurements. M.S.A., J.R.W. and G.S.S. were responsible for the synthetic biology. D.W.A., R.D.D., C.D.C., J.M.M. and M.L.S. analysed the experimental data. J.M.M., N.F.S. and C.D.C. were responsible for simulations. C.D.C. and M.L.S. developed the frequency domain analytical approach. M.L.S. was responsible for integration of experimental, analytical and computational components.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Video
This movie shows fluorescence time-lapsed microscopy of one experiment using the pGFPasv gene circuit. The time lapse covers 8 hours with frames separated by 5 minutes. The yellow outline around one cell follows a single trajectory through six generations of cell division. The cell doubling time was approximately 90 minutes. (MOV 973 kb)
Supplementary Methods
Detailed methods are described here for the genetic constructs and cell strains; the extraction of noise frequency composition from fluorescence time-lapsed microscopy; and the analytical and computational modeling of the gene circuit noise properties. (DOC 2113 kb)
Rights and permissions
About this article
Cite this article
Austin, D., Allen, M., McCollum, J. et al. Gene network shaping of inherent noise spectra. Nature 439, 608–611 (2006). https://doi.org/10.1038/nature04194
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04194
This article is cited by
-
Negative autoregulation controls size scaling in confined gene expression reactions
Scientific Reports (2022)
-
The emergence of the two cell fates and their associated switching for a negative auto-regulating gene
BMC Biology (2019)
-
Stem cell bioengineering: building from stem cell biology
Nature Reviews Genetics (2018)
-
Stochastic gene expression in Arabidopsis thaliana
Nature Communications (2017)
-
Feedback Regulation and Its Efficiency in Biochemical Networks
Journal of Statistical Physics (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.